For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVGADFTAHAEPYRRELLAHCYRMTGSLHDAEDLVQETLLRAWKAYDRFEGKSSVRTWLHRIATNTCLSA LEGRQRRPLPVGLGAPSADPTAELVERREVPWLEPLPALSDDPADPSVIVGSRESVRLAFVAALQYLSPR QRAVLLLRDVLGWRAAEVAEAIGTTTAAVNSLLQRARAQLDAVGPSADDQPAQPDSAETQDLLARYIAAF ESYDIDRLVELFTAEAIWEMPPYTGWYQGARTIVTLIHQQCPAEGAGDMRLLPLIANGQPAAAMYMRAGD VHLPFQLHVLDVRGDRVSHVVAFLDDSLFAKFGLPAALGPRQEGVRA
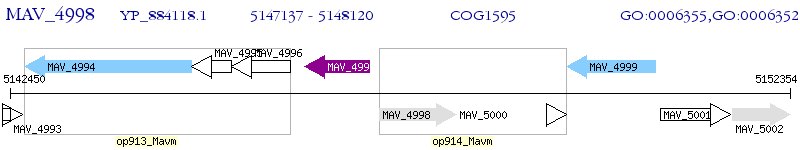
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4998 | - | - | 100% (327) | RNA polymerase factor sigma-70 |
| M. avium 104 | MAV_1129 | - | 7e-21 | 29.08% (282) | sigma factor |
| M. avium 104 | MAV_2018 | - | 8e-19 | 28.66% (321) | RNA polymerase ECF-subfamily protein sigma factor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0188c | sigG | 1e-152 | 83.44% (314) | RNA polymerase factor sigma-70 |
| M. gilvum PYR-GCK | Mflv_0519 | - | 1e-128 | 71.20% (316) | RNA polymerase factor sigma-70 |
| M. tuberculosis H37Rv | Rv0182c | sigG | 1e-152 | 83.44% (314) | RNA polymerase factor sigma-70 |
| M. leprae Br4923 | MLBr_01076 | sigE | 8e-12 | 30.82% (159) | RNA polymerase sigma factor SigE |
| M. abscessus ATCC 19977 | MAB_4553 | - | 1e-123 | 69.21% (315) | RNA polymerase factor sigma-70 |
| M. marinum M | MMAR_0426 | sigG | 1e-151 | 82.59% (316) | alternative RNA polymerase sigma factor SigG |
| M. smegmatis MC2 155 | MSMEG_0219 | - | 1e-125 | 70.48% (315) | RNA polymerase factor sigma-70 |
| M. thermoresistible (build 8) | TH_0604 | sigG | 1e-126 | 71.79% (319) | PROBABLE ALTERNATIVE RNA POLYMERASE SIGMA FACTOR SIGG (RNA |
| M. ulcerans Agy99 | MUL_1076 | sigG | 1e-149 | 81.65% (316) | RNA polymerase factor sigma-70 |
| M. vanbaalenii PYR-1 | Mvan_0149 | - | 1e-122 | 68.99% (316) | RNA polymerase factor sigma-70 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0426|M.marinum_M -----------------------MSALAGNSTQNSIANFTAPTAERERPA
MUL_1076|M.ulcerans_Agy99 -------------MSATLRRLIDVSALAGNSTQNSIANFTAPTAERERPA
MAV_4998|M.avium_104 ------------------------------------------------MV
Mb0188c|M.bovis_AF2122/97 MRTSPMPAKFRSVRVVVITGSVTAAPVRVSETLRRLIDVSVLAENSGREP
Rv0182c|M.tuberculosis_H37Rv MRTSPMPAKFRSVRVVVITGSVTAAPVRVSETLRRLIDVSVLAENSGREP
MAB_4553|M.abscessus_ATCC_1997 --------------------------------------------MTAHAI
MSMEG_0219|M.smegmatis_MC2_155 ----------------------------MSVILRKLTTVTVLAHKLDDDS
TH_0604|M.thermoresistible__bu ----------------------------MSVMLRTLLHVTVLTAPPDGGS
Mflv_0519|M.gilvum_PYR-GCK ----------------------------MSHPMRKLLFVTVLA--LDEGG
Mvan_0149|M.vanbaalenii_PYR-1 ----------------------------MSQPVRKLHFVTVTA--LDDSS
MLBr_01076|M.leprae_Br4923 ---------------------------------------------MGDGI
MMAR_0426|M.marinum_M GD----EFAVHAEPYRRELLAHCYRMTGSVHDAEDLVQETLLRAWKSYHR
MUL_1076|M.ulcerans_Agy99 GD----EFAVHAEPYRRELLAHCYRMTGSVHDAEDLVQETLLRAWKSYHR
MAV_4998|M.avium_104 GA----DFTAHAEPYRRELLAHCYRMTGSLHDAEDLVQETLLRAWKAYDR
Mb0188c|M.bovis_AF2122/97 ADERRGDFSAHTEPYRRELLAHCYRMTGSLHDAEDLVQETLLRAWKAYEG
Rv0182c|M.tuberculosis_H37Rv ADERRGDFSAHTEPYRRELLAHCYRMTGSLHDAEDLVQETLLRAWKAYEG
MAB_4553|M.abscessus_ATCC_1997 ED----EFLSHTEQYRREILAHCYRMTGSIHDAEDLVQETYLRGWKAYAR
MSMEG_0219|M.smegmatis_MC2_155 PA---DAFLAEAQTYRRELLAHCYRMTGSLHDAEDLVQETYLRAWKSYQG
TH_0604|M.thermoresistible__bu -----DAFLADAEQYRRELLAHCYRMTGSLHDAEDLVQETFLRAWKSHHK
Mflv_0519|M.gilvum_PYR-GCK AK---DAFLADAQKYRRELLAHCYRMTGSLHDAEDLVQETYLRAWKSYDR
Mvan_0149|M.vanbaalenii_PYR-1 AE---DAFLADAQRYRRELLAHCYRMTGSLHDAEDLVQETYLRAWKSFKG
MLBr_01076|M.leprae_Br4923 GMEREGRWTGNTQCPLRVVPGDESPTLDGRASPEDLIITNLLSPTIMSHP
: .:: * : .. .. ..***: . *
MMAR_0426|M.marinum_M FEGKSSLRTWLHRIATNTCLSALEGRQRRPLPTGLAASGSDPTAPLVERA
MUL_1076|M.ulcerans_Agy99 FEGKSSLRTWLHRIATNTCMSALEGRQRRPLPTGLAASGSDPTAPLVERA
MAV_4998|M.avium_104 FEGKSSVRTWLHRIATNTCLSALEGRQRRPLPVGLGAPSADPTAELVERR
Mb0188c|M.bovis_AF2122/97 FAGKSSLRTWLHRIATNTCLTALEGRRRRPLPTGLGRPSADPSGELVERR
Rv0182c|M.tuberculosis_H37Rv FAGKSSLRTWLHRIATNTCLTALEGRRRRPLPTGLGRPSADPSGELVERR
MAB_4553|M.abscessus_ATCC_1997 FEGKSSVRTWLYRIATNTCLTALEGKQRRPLPTGLGAPSADPLDELDERS
MSMEG_0219|M.smegmatis_MC2_155 FQGRSSVRTWLYRIATNTCLTSLEGRSRRPLPSGLGTPSSAPTDQITEHH
TH_0604|M.thermoresistible__bu FEGKASVRTWLYKIATNACLTALDGRQRRPLPTGLGAPSSDPTDDIVARE
Mflv_0519|M.gilvum_PYR-GCK FEGKSSVRTWLYRIATNTCLTALDGRKRRPLPSGLGGPSADPYADLTERH
Mvan_0149|M.vanbaalenii_PYR-1 FQGKSSVRTWLYRIATNTCLTALDGNKRRALPSGLGQPASDPAGELFVRP
MLBr_01076|M.leprae_Br4923 PPSRD-----------DDWVEPFDALQGTAVFDATGDKATMPSWEELVRQ
.: : : .::. .: . . .: * :
MMAR_0426|M.marinum_M EVPWLEPLPDL-MADPADPSVIVGSRESVRLAFVAALQHLSPRQRAVLLL
MUL_1076|M.ulcerans_Agy99 EVPWLEPLPDL-MADPADPSVIVGSRESVRLAFVAALQHLSPRQRAVLLL
MAV_4998|M.avium_104 EVPWLEPLPAL-SDDPADPSVIVGSRESVRLAFVAALQYLSPRQRAVLLL
Mb0188c|M.bovis_AF2122/97 EVSWLEPLPDV-TDDPADPSTIVGNRESVRLAFVAALQHLSPRQRAVLLL
Rv0182c|M.tuberculosis_H37Rv EVSWLEPLPDV-TDDPADPSTIVGNRESVRLAFVAALQHLSPRQRAVLLL
MAB_4553|M.abscessus_ATCC_1997 EIPWLEPLP----DDATDPASIVGARESVRLAFVAALQHLPAKQRAVLVL
MSMEG_0219|M.smegmatis_MC2_155 EVPWLEPLPDG--DDPADPSQIVGTRESVRLAFVAALQHLSPRQRAVLVM
TH_0604|M.thermoresistible__bu EVPWLEPLP----DDAADPSTIVGSRESVRLAFVAALQHLPPRQRAVLLL
Mflv_0519|M.gilvum_PYR-GCK EIAWLEPIPDDPHEDPADPSVIAESRESVRLALIAALQHLSPRQRAVLVL
Mvan_0149|M.vanbaalenii_PYR-1 EVTWLEPLPDAPREDPSDPSVIAESRESVRLAFIAALQHLPPRQRAVLVL
MLBr_01076|M.leprae_Br4923 HAARVYWLAYRLSGNQHDAEDLT-------------------QETFIRVF
. . : :. : *. :. :: : ::
MMAR_0426|M.marinum_M RDVLQWKAAEVAEAIGTTTVAVNSLLQRARAQLETAQPSADDGLVAPDSP
MUL_1076|M.ulcerans_Agy99 RDVLQWKAAEVAQAIGTTTVAVNSLLQRARAQLETVQPSADDGLVAPDSP
MAV_4998|M.avium_104 RDVLGWRAAEVAEAIGTTTAAVNSLLQRARAQLDAVGPSADDQPAQPDSA
Mb0188c|M.bovis_AF2122/97 RDVLQWKSAEVADAIGTSTVAVNSLLQRARSQLQTVRPSAADRLSAPDSP
Rv0182c|M.tuberculosis_H37Rv RDVLQWKSAEVADAIGTSTVAVNSLLQRARSQLQTVRPSAADRLSAPDSP
MAB_4553|M.abscessus_ATCC_1997 REVLQWKAQEVAEAIGSTTAAVNSLLQRARAQLHEVSP-HEETLLEPEDP
MSMEG_0219|M.smegmatis_MC2_155 REVLQWKASEVGEAIGASTAAVNSLLQRARAQLEAVGPNEDDKIEAPDSP
TH_0604|M.thermoresistible__bu REVLQWRAAEVAEVLNTSTAAVNSLLQRARAQLDQVGPSEDDELVPPESP
Mflv_0519|M.gilvum_PYR-GCK REVLQWKAAEVGEVIGASTAAVNSLLQRARAQLDEVAPSRDDQPVSPESP
Mvan_0149|M.vanbaalenii_PYR-1 REVLQWKAAEVGEAVGTSTAAVNSLLQRARAQLDEISPSRDDEPVPPESP
MLBr_01076|M.leprae_Br4923 RSVQNYQPGTFEGWLHRITTNLFLDMVRRRARIRMDALPDDYDRVPADEP
*.* ::. . : *. : : * *::: .:..
MMAR_0426|M.marinum_M EAQDLLTRYITAFEAYDIDRLVEIFTAEAIWEMPPYAGWYQGARDIITLI
MUL_1076|M.ulcerans_Agy99 EAQDLLTRYIAAFEAHDIDRLVEIFTAEAIWEMPPYAGWYQGARDIITLI
MAV_4998|M.avium_104 ETQDLLARYIAAFESYDIDRLVELFTAEAIWEMPPYTGWYQGARTIVTLI
Mb0188c|M.bovis_AF2122/97 EAQDLLARYIAAFEAYDIDRLVELFTAEAIWEMPPYTGWYQGAQAIVTLI
Rv0182c|M.tuberculosis_H37Rv EAQDLLARYIAAFEAYDIDRLVELFTAEAIWEMPPYTGWYQGAQAIVTLI
MAB_4553|M.abscessus_ATCC_1997 MLREQLRQYINAFERYDVEAIAKMFTTEAIWEMPPFTGWYQGGPSIAALI
MSMEG_0219|M.smegmatis_MC2_155 QAQATLDRYIAAFEAYDIDKLVELFTEEAIWEMPPFDSWYQGPQAIGDLS
TH_0604|M.thermoresistible__bu EAQELLTRYIDAFESYDIDKLVELFTADAVWEMPPFDGWYQGPQNIVTLS
Mflv_0519|M.gilvum_PYR-GCK QAAEALAKYIAAFEDYDMDRLVELFTADAVWEMPPFDGWYSGPADIIALS
Mvan_0149|M.vanbaalenii_PYR-1 EAAELLDKYIAAFEDYDMDRLVELFTDDAVWEMPPFDGWYQGPANIVTLS
MLBr_01076|M.leprae_Br4923 NPEQIYH-----------DSRLGPDLQAALDSLPPEFRAAVVLCDIEGLS
: *: .:** * *
MMAR_0426|M.marinum_M HQQCPAECPGDMRLIPLVAN-GQPAAAMYMRAGE--THVPFQLHVLDMVD
MUL_1076|M.ulcerans_Agy99 HQQCPAECPGDMRLIPLVAN-GQPAAAMYMRAGE--TRVPFLLHVLDMVE
MAV_4998|M.avium_104 HQQCPAEGAGDMRLLPLIAN-GQPAAAMYMRAGD--VHLPFQLHVLDVRG
Mb0188c|M.bovis_AF2122/97 HQQCPAYSPGDMRLISLIAN-GQPAAAMYMRAGD--VHLPFQLHVLDMAA
Rv0182c|M.tuberculosis_H37Rv HQQCPAYSPGDMRLISLIAN-GQPAAAMYMRAGD--VHLPFQLHVLDMAA
MAB_4553|M.abscessus_ATCC_1997 KEQCPAEHAGDMRLVETSAN-GQPAIGLYMREGDG-VHRPFQMHVLDVTQ
MSMEG_0219|M.smegmatis_MC2_155 RYKCPAEEPGDMRFLQTTAN-GQPAAAMYMLNRETGRHEAFQLHVLDITA
TH_0604|M.thermoresistible__bu KTHCPAERPGDMRLIPTTAN-GQPAGAMYLRNPETGTHEAFQMHVLAIGP
Mflv_0519|M.gilvum_PYR-GCK KYHCPAAGAGDMRFLRTTAN-GQPVAALYMRNPETNVHEAFQLHVLDLRT
Mvan_0149|M.vanbaalenii_PYR-1 KVQCPAEKAGDMRFLRTTAN-GQPVAALYMRNPETGVHEAFQLHVLDAGK
MLBr_01076|M.leprae_Br4923 YEEIGATLGVKLGTVRSRIHRGRQALRDYL----------LTAHPRDGAT
. * .: : : *: . *: : *
MMAR_0426|M.marinum_M GRVSHVTAFLDGSLFPKFGLPASI---------
MUL_1076|M.ulcerans_Agy99 GRVSHVTAFLDGSLFPKFGLPASI---------
MAV_4998|M.avium_104 DRVSHVVAFLDDSLFAKFGLPAALGPRQEGVRA
Mb0188c|M.bovis_AF2122/97 DRVSHVVAFLDTTLFPKFGLPDSL---------
Rv0182c|M.tuberculosis_H37Rv DRVSHVVAFLDTTLFPKFGLPDSL---------
MAB_4553|M.abscessus_ATCC_1997 DGVAHVVCFFDISLFEKFGLPAELTD-------
MSMEG_0219|M.smegmatis_MC2_155 DGISHVVAFMGEGLFKKFGLPETL---------
TH_0604|M.thermoresistible__bu GGISHVVAFHMP-SFAPFGLPDSL---------
Mflv_0519|M.gilvum_PYR-GCK RGIAHVVAFMDVSLFRKFGLPETL---------
Mvan_0149|M.vanbaalenii_PYR-1 AGITHVVAFKENDLFARFGLPDTL---------
MLBr_01076|M.leprae_Br4923 TDATHVKSA------------------------
:** .