For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NLRVTVFGATGQIGRFVVADLLADGHAATAYVRNPGKLQVADPHLTVATGELSDAEAVRKAVRGADAVIS ALGPSLSRRAKGTPVTEGTRNIVAAMQAEHVSRYIGLATPSVPDSRDRPTLKAKILPIIAGTLFPNALGE IVGMTKAVTDSDLAWTIARITSPNNSRPKGTLRVGFLGRDKVGSVMSRADIAAFLVAQLDDETFIRAAPA ISN*
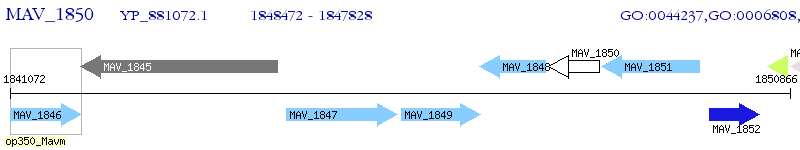
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1850 | - | - | 100% (214) | hypothetical protein MAV_1850 |
| M. avium 104 | MAV_1225 | - | 2e-08 | 32.79% (122) | 3-beta hydroxysteroid dehydrogenase/isomerase family protein |
| M. avium 104 | MAV_2451 | - | 1e-07 | 30.40% (125) | hypothetical protein MAV_2451 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3752 | - | 5e-08 | 29.93% (147) | putative oxidoreductase |
| M. gilvum PYR-GCK | Mflv_1635 | - | 7e-07 | 30.91% (110) | NAD-dependent epimerase/dehydratase |
| M. tuberculosis H37Rv | Rv3725 | - | 5e-08 | 29.93% (147) | oxidoreductase |
| M. leprae Br4923 | MLBr_01942 | - | 1e-05 | 30.00% (120) | putative cholesterol dehydrogenase |
| M. abscessus ATCC 19977 | MAB_3697 | - | 2e-09 | 26.24% (221) | putative epimerase/dehydratase |
| M. marinum M | MMAR_5252 | - | 8e-08 | 30.82% (146) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_5228 | - | 4e-08 | 33.33% (120) | 3-beta hydroxysteroid dehydrogenase/isomerase family protein |
| M. thermoresistible (build 8) | TH_3137 | - | 3e-19 | 36.02% (161) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_4328 | - | 5e-08 | 30.82% (146) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_5113 | - | 4e-08 | 32.41% (108) | NAD-dependent epimerase/dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3752|M.bovis_AF2122/97 ---------MQNATMRVLVTGGTGFVGGWTAKAIADAGHSVRFLVRNPAR
Rv3725|M.tuberculosis_H37Rv ---------MQNATMRVLVTGGTGFVGGWTAKAIADAGHSVRFLVRNPAR
MMAR_5252|M.marinum_M ---------MHNRCMHVLVTGGTGFVGGWTAKAIADAGHSVRFLVRNPAK
MUL_4328|M.ulcerans_Agy99 ---------MHNRCMHVLVTGGTGFVGGWTAKAIADAGHSVRFLVRNPAK
Mflv_1635|M.gilvum_PYR-GCK ---------MPGNDVRCVVTGATGYIGGRLVPELLARGLQVRAMARTPSK
Mvan_5113|M.vanbaalenii_PYR-1 ---------MSTEQVRCLVTGATGYIGRRLVPELLDRGHTVRAMARTPAK
MLBr_01942|M.leprae_Br4923 MLRGVGNASLTTELGRVLVTGGSGFVGANLVTALLERGYQVRSFDRAPMP
MSMEG_5228|M.smegmatis_MC2_155 ------MADSTTDLGRILVTGGSGFVGANLVTELLDRGYAVRSFDRVPSP
MAV_1850|M.avium_104 ------------MNLRVTVFGATGQIGRFVVADLLADGHAATAYVRNPGK
TH_3137|M.thermoresistible__bu --------------MRIVIFGANGPTGRLLTTRVLDAGHQAVAVTRRPTD
MAB_3697|M.abscessus_ATCC_1997 --------------MKIAIFGATGMVGSRIAAELRSRGHQVTGYSRRGS-
: : *..* * . : * . *
Mb3752|M.bovis_AF2122/97 LKTSVAKLGVDVSDFAVADISDRDSVREALNGCDAVVHSAALVATD----
Rv3725|M.tuberculosis_H37Rv LKTSVAKLGVDVSDFAVADISDRDSVREALNGCDAVVHSAALVATD----
MMAR_5252|M.marinum_M LESSVAKLGVDVSDFIIGDIKDRDLVREALTGCDAVVHSAALVATD----
MUL_4328|M.ulcerans_Agy99 LESSVAKLGVDVSDFIIGDIKDRDLVREALTGCDAVVHSAALVATD----
Mflv_1635|M.gilvum_PYR-GCK LDRTEWRD---RVEVVRGDLTEADSLTAAFNGADVVYY---LVHSMGTSR
Mvan_5113|M.vanbaalenii_PYR-1 LDDAAWRD---RVEVVRGDLTEPDSLASAFDGVDVIYY---LVHSMGTSR
MLBr_01942|M.leprae_Br4923 LPQHPQ------LEVLQGDITDATVCTTAMDSIDTVFHTAAIIELMGGAS
MSMEG_5228|M.smegmatis_MC2_155 LPAHAG------LEVATGDICDLDNVTNAVAGVDTVFHTAAIIDLMGGAS
MAV_1850|M.avium_104 LQVADP-----HLTVATGELSDAEAVRKAVRGADAVISALGPSLSR----
TH_3137|M.thermoresistible__bu FPMRHP-----RLTVAGADVRDPATLDGVVHGADAVLSSLGVPFSM----
MAB_3697|M.abscessus_ATCC_1997 ------------DSTRAGTLTDGALVATVAAGHDAVVSAIGPSRSG----
. : : . . *.:
Mb3752|M.bovis_AF2122/97 --PRETSRMLSTNMAGAQNVLGQAVELGMDPIVHVSSFTALFRPNLATLS
Rv3725|M.tuberculosis_H37Rv --PRETSRMLSTNMAGAQNVLGQAVELGMDPIVHVSSFTALFRPNLATLS
MMAR_5252|M.marinum_M --QRQTQDMLSTNMEGARNVLGQAVALGLDPIVHVSSFTALFHPGLQTLS
MUL_4328|M.ulcerans_Agy99 --QRQTQDMLSTNMEGARNVLGQAVALGLDPIVHVSSFTALFHPGLQTLS
Mflv_1635|M.gilvum_PYR-GCK DFVAEERR-------SAHNVVEAAKKAGVRRIVYLSG-------------
Mvan_5113|M.vanbaalenii_PYR-1 DFVAEEAR-------SARNVVAAAEKAGVRRLVYLGG-------------
MLBr_01942|M.leprae_Br4923 VTDEYRQRSYTVNVGGTENLLRAGQKSGVKRFVYTASNSVVMGGTPITGG
MSMEG_5228|M.smegmatis_MC2_155 VTAEYRQRSFAVNVGGTENLVRAAQSAGVKRFVYTASNSVVMGGQHIVHG
MAV_1850|M.avium_104 ------RAKGTPVTEGTRNIVAAMQAEHVSRYIGLAT-------------
TH_3137|M.thermoresistible__bu -------RAIDTYSVGTANIVAAMRAAAVPRLVVTSSTG-----------
MAB_3697|M.abscessus_ATCC_1997 ----------EPHSEFIEAITAAAANLHGTRLFVVGG-------------
: . .
Mb3752|M.bovis_AF2122/97 ADLPVAGGTDGYGQSKAQIEIYARGLQDAG------APVNITYPGMVLGP
Rv3725|M.tuberculosis_H37Rv ADLPVAGGTDGYGQSKAQIEIYARGLQDAG------APVNITYPGMVLGP
MMAR_5252|M.marinum_M ADLPVVGGSDGYGTSKAQVEIYARGLQDAG------APVNITYPGMVLGP
MUL_4328|M.ulcerans_Agy99 ADLPVVGGSDGYGTSKAQVEIYARGLQDAG------APVNITYPGMVLGP
Mflv_1635|M.gilvum_PYR-GCK --LHPD-RKELSRHLASRVEVGEILMQSG-------IETVVLQAGIVIGS
Mvan_5113|M.vanbaalenii_PYR-1 --LHPQGKTDLSRHLASRVEVGDILLGST-------VETVVLQAGIVVGS
MLBr_01942|M.leprae_Br4923 DETMPYTKRFNDLYTETKVVAEKFVLSQNGVPDGETMLTCSIRPSGIWGR
MSMEG_5228|M.smegmatis_MC2_155 DETLPYTERFNDLYTETKVVAEKFVLGRNGVAG---MLTCSIRPSGIWGR
MAV_1850|M.avium_104 ---PSVPDSRDRPTLKAKILP---------------IIAGTLFPN-----
TH_3137|M.thermoresistible__bu --AFPYPNRVDPPFALRIVEP---------------VITRTIGKSTYDDQ
MAB_3697|M.abscessus_ATCC_1997 --AGTLRVNGVRLVDSP---------------------------------
Mb3752|M.bovis_AF2122/97 PVGDQFGEAGEGVRSALWMHVIPGR----GAAWLIVDVRDVAALHAALLE
Rv3725|M.tuberculosis_H37Rv PVGDQFGEAGEGVRSALWMHVIPGR----GAAWLIVDVRDVAALHAALLE
MMAR_5252|M.marinum_M PVGNQFGEAGEGVKAALEIRTIPGR----NAAWLIIDVRDVAAVHAALLE
MUL_4328|M.ulcerans_Agy99 PVGNQFGEAGEGVKAALEIRTIPGR----NAAWLIIDVRDVAAVHAALLE
Mflv_1635|M.gilvum_PYR-GCK GS-----ASFEMVRHLTDRLPVMTTPKWVHNKIQPISISDVLHYLAEAAT
Mvan_5113|M.vanbaalenii_PYR-1 GS-----ASFEMVRHLTDRLPVMTTPKWVHNTIQPIAIADVLYYLAEAAT
MLBr_01942|M.leprae_Br4923 GDQTMFRKAFESVVSGHVKVLIGSKNAKLDNSYVHNLVHGLILAAEHLVP
MSMEG_5228|M.smegmatis_MC2_155 GDQTMFRKVFESVLAGHVKVLVGRKSTLLDNSYVHNLVHGFILAAEHLTP
MAV_1850|M.avium_104 -------ALGEIVG-----------------MTKAVTDSDLAWTIARITS
TH_3137|M.thermoresistible__bu R------RMESIVRGSGLDWTIVRP------SGLFDLAEPTDYVAGEVEP
MAB_3697|M.abscessus_ATCC_1997 -------EFPEPYRAEALAHAAALD-------TLEQTAQDVDWVYLSPAP
.
Mb3752|M.bovis_AF2122/97 SGRGPRR--YTAGGHRIPVPELAKILGEVAGTTMLAVPVPDSALRVAGSV
Rv3725|M.tuberculosis_H37Rv SGRGPRR--YTAGGHRIPVPELAKILGGSPAPRCWPSRCPIPRCVSRDRC
MMAR_5252|M.marinum_M PGRGPRR--YMVGGHRIPVPELAKMLGQVAETPIVSVPIPDSVLRVAGSL
MUL_4328|M.ulcerans_Agy99 PGRGPRR--YMVGGHRIPVPELAKMLGQVAETPIVSVPIPDSVLRVAGSL
Mflv_1635|M.gilvum_PYR-GCK AEFGESRTWDIGGPDVMEYGDAMQGYADVAGLRRRVMVVLPFLTPTIASW
Mvan_5113|M.vanbaalenii_PYR-1 AEVPESRTWDIGGPDVMEYGEAMQIYADIAHLRRRVMVVLPFLTPTIASL
MLBr_01942|M.leprae_Br4923 GGTAPGQAYFINDGEPINFFDFMGPIIKACGENWPRVRISGRLVRNVMAV
MSMEG_5228|M.smegmatis_MC2_155 NGTAPGQAYFINDGEPVNMFEFARPVIEACGRKLPRVRVPGRAVHAAMSG
MAV_1850|M.avium_104 PNNSRPK-----GTLRVGFLGRDKVGSVMSRADIAAFLVAQLDDETFIRA
TH_3137|M.thermoresistible__bu VGMFTAR------IDLADYMTRLAGSSDGTGKTVVVSTVEHIPTIWQMVR
MAB_3697|M.abscessus_ATCC_1997 LIEPGKR------TGSYRVAGDAPAGQSISAEDYAVAVADELEHPAHRRE
:
Mb3752|M.bovis_AF2122/97 LDQAGPYLPFNTPFTAAGMQYYTQMPEPD----DSPSEKELGITYRDPRD
Rv3725|M.tuberculosis_H37Rv WIKPGP-------------ICLSILRSPR----QVCS-----TTHRCRSP
MMAR_5252|M.marinum_M LDLAGPYLPFDNPFTAAGMQYYTQMPDSD----DSPSELELGVTYRDPRT
MUL_4328|M.ulcerans_Agy99 LDLAGPYLPFDNPFTAAGMQYYTQMPDSD----DSPSELELGVTYRDPRT
Mflv_1635|M.gilvum_PYR-GCK WVGLVTPIPSGLARPLVESLECDAICHEHDIDAVIPPPKGGLIGYREAVA
Mvan_5113|M.vanbaalenii_PYR-1 WVGLVTPIPSGLARPLVESLECDAVAHEHDIDTVIPPPEGGVTGYRDAVT
MLBr_01942|M.leprae_Br4923 WQRLHFGF--GLPKPPMEPLAVERVYLDNYFSIEKAHKE---LGYRPLFT
MSMEG_5228|M.smegmatis_MC2_155 WQRLHFRF--GIPEPLLEPLAVERLYLNNYFSIAKATRD---LGYRPLFT
MAV_1850|M.avium_104 APAISN--------------------------------------------
TH_3137|M.thermoresistible__bu REAVKTG-------------------------------------------
MAB_3697|M.abscessus_ATCC_1997 RFTVAN--------------------------------------------
Mb3752|M.bovis_AF2122/97 TVADTVTALRGLGS-----------------------
Rv3725|M.tuberculosis_H37Rv TIRRAKKN-----------------------------
MMAR_5252|M.marinum_M TLADTVAALTA--------------------------
MUL_4328|M.ulcerans_Agy99 TLADTVAALTA--------------------------
Mflv_1635|M.gilvum_PYR-GCK KELRSEGGSS-AGRRHLLRFSPAASD-----------
Mvan_5113|M.vanbaalenii_PYR-1 QALRDDGTAAKGGRRHLLRFSKVVPQ-----------
MLBr_01942|M.leprae_Br4923 TEQAMAECLPYYTELFEQIKVAAKPHLASIAATPRPE
MSMEG_5228|M.smegmatis_MC2_155 TEQARVDCLPYYVDLFKQMEAQARPA-----------
MAV_1850|M.avium_104 -------------------------------------
TH_3137|M.thermoresistible__bu -------------------------------------
MAB_3697|M.abscessus_ATCC_1997 -------------------------------------