For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNFTSKVAVVTGAASGIGEATAKRLAEDGAKVVVADINEDGGERVVAEIAERGGEAAFLRTDISIEEDVN SLIDFAVSTYGGLHLAHNNAAVAHTPTPLHELSTDVFDTVTGIGLRGTFFCMRAEIAHMIGHGGGAIVNT ASGAALKAAAGMHAYVATKHGIVGLTRNAGLDYARNGIRVNAVAPGSIKTPQMASFPQEMQDEWERIIPM GRMGRPEEVADVVAFLLSDAASFITATTVEIDGAYLQSSK
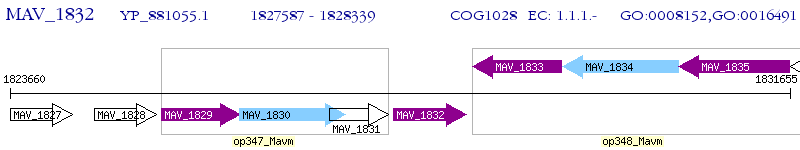
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1832 | - | - | 100% (250) | 2,5-dichloro-2,5-cyclohexadiene-1,4-diol dehydrogenase |
| M. avium 104 | MAV_3714 | - | 7e-43 | 39.51% (243) | short chain dehydrogenase |
| M. avium 104 | MAV_0886 | - | 2e-42 | 37.55% (253) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2882c | - | 4e-42 | 38.93% (244) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4062 | - | 8e-46 | 41.56% (243) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv2857c | - | 4e-42 | 38.93% (244) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_02565 | fabG | 7e-23 | 29.10% (244) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. abscessus ATCC 19977 | MAB_1219 | - | 2e-38 | 38.87% (247) | short chain dehydrogenase |
| M. marinum M | MMAR_4765 | - | 9e-41 | 37.01% (254) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2598 | - | 6e-44 | 42.21% (244) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_3693 | - | 4e-40 | 39.08% (238) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_0336 | - | 1e-40 | 35.83% (254) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_2281 | - | 3e-44 | 40.33% (243) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_1832|M.avium_104 --------------------------------------------------
MAB_1219|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_2598|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4062|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2281|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4765|M.marinum_M --------------------------------------------------
MUL_0336|M.ulcerans_Agy99 --------------------------------------------------
TH_3693|M.thermoresistible__bu --------------------------------------------------
MLBr_02565|M.leprae_Br4923 MAPKCSSDLFSQIVSSGPGSFVAKQLGVPRPETLRRYRPSDAPLCGSLLI
MAV_1832|M.avium_104 --------------------------------------------------
MAB_1219|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_2598|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4062|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2281|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4765|M.marinum_M --------------------------------------------------
MUL_0336|M.ulcerans_Agy99 --------------------------------------------------
TH_3693|M.thermoresistible__bu --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GGDGRLVEPLRATLEKDYDLVSNNLGGRWTDSFGGLVFDATGITAPAELK
MAV_1832|M.avium_104 --------------------------------------------------
MAB_1219|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_2598|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4062|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2281|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4765|M.marinum_M --------------------------------------------------
MUL_0336|M.ulcerans_Agy99 --------------------------------------------------
TH_3693|M.thermoresistible__bu --------------------------------------------------
MLBr_02565|M.leprae_Br4923 GLHNFFTPLLRNLGRCARVVVIGTTPDATTSTDERIAQRALEGFSRSLGK
MAV_1832|M.avium_104 --------------------------------------------------
MAB_1219|M.abscessus_ATCC_1997 --------------------------------------------------
Mb2882c|M.bovis_AF2122/97 --------------------------------------------------
Rv2857c|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_2598|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_4062|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2281|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_4765|M.marinum_M --------------------------------------------------
MUL_0336|M.ulcerans_Agy99 --------------------------------------------------
TH_3693|M.thermoresistible__bu --------------------------------------------------
MLBr_02565|M.leprae_Br4923 ELRRGATVALVYLSPDAKPAATGLESTMRFILSAKSAYVNGQVFYVGAAD
MAV_1832|M.avium_104 --------MNFTSKVAVVTGAASGIGEATAKRLAEDGAKVVVADIN--ED
MAB_1219|M.abscessus_ATCC_1997 -------MGEFDNKVAIVTGAAQGIGEAYAHALAREGAAVVVADIN--AE
Mb2882c|M.bovis_AF2122/97 ---MMDLSQRLAGRVAVITGGGSGIGLAAGRRMRAEGATIVVGDVD--VE
Rv2857c|M.tuberculosis_H37Rv ---MMDLSQRLAGRVAVITGGGSGIGLAAGRRMRAEGATIVVGDVD--VE
MSMEG_2598|M.smegmatis_MC2_155 ----MDLTQRLAGKVAVITGGASGIGLATGRRLRAEGATVVVGDID--PT
Mflv_4062|M.gilvum_PYR-GCK ----MDLSQRLKDKVAVITGGASGIGLATAKRMRSEGAIVVIGDLD--EK
Mvan_2281|M.vanbaalenii_PYR-1 ----MDLGQRLKDKVAVITGGASGIGLATAKRMQAEGAIVVIGDVD--AS
MMAR_4765|M.marinum_M ------MVDELAGKIAIVTGGASGIGRATVARFIAEGARVVIADVE--EE
MUL_0336|M.ulcerans_Agy99 ------MVDELAGKIAIVTGGASGIGRATVARFIAEGARVVIADVE--EE
TH_3693|M.thermoresistible__bu -------VSLLAGQTAVVTGGARGLGYAIAERFIAEGARVVIGDID--ES
MLBr_02565|M.leprae_Br4923 STPPTNWDRPLENKVAIVTGAARGIGAAIAEVVARDGARVVAIDVESAAE
: .: *::**.. *:* * . :** :* *::
MAV_1832|M.avium_104 GGERVVAEIAERGGEAAFLRTDISIEEDVNSLIDFAVSTYG-GLHLAHNN
MAB_1219|M.abscessus_ATCC_1997 MGEGVAKQIVADGGRAIFAGVDVSDPDSAIAMADKAVSEFG-GIDYLVNN
Mb2882c|M.bovis_AF2122/97 AGGAAADELS-----GLFVPTDVCDEDAVNGLFDGAAETYG-RIDIAFNN
Rv2857c|M.tuberculosis_H37Rv AGGAAADELS-----GLFVPTDVCDEDAVNGLFDGAAETYG-RIDIAFNN
MSMEG_2598|M.smegmatis_MC2_155 TGKAAADELE-----GLFVPVDVSEQEAVDNLFDTAASTFG-RVDIAFNN
Mflv_4062|M.gilvum_PYR-GCK TGKSVANDLN-----VTFVQVDVSDQVAVDNLFDTAFEVHG-GVDIAFNN
Mvan_2281|M.vanbaalenii_PYR-1 TGKSVANDMN-----VTFVQVDVADQVAVDHLFDTAFEVHG-GVDIAFNN
MMAR_4765|M.marinum_M RGESLAAALG--ADAMF-CRTDVSQPEQVAAVVAAAVENFG-GLHVMVNN
MUL_0336|M.ulcerans_Agy99 RGESLAAALG--ADAMF-CRTDVSQPEQVAAVVAAAVDNFG-GLHVMVNN
TH_3693|M.thermoresistible__bu ATRAAADSLGG-ADVAVGMRCDVTRSDEVEALVGAAVKRFG-SLDVMVNN
MLBr_02565|M.leprae_Br4923 ALDETANRVG-----GTTLSLDVTADDAVDKITEHLHDYQGGHADILVNN
. : *: . : . * . **
MAV_1832|M.avium_104 AAVAH--TPTPLHELSTDVFDTVTGIGLRGTFFCMRAEIAHMIGHGGGAI
MAB_1219|M.abscessus_ATCC_1997 AAIYGGMKLDLLLSVPWDYYKKFMSVNQDGALVCTRAVYKHIAQRGGGAI
Mb2882c|M.bovis_AF2122/97 AGISPP-EDNLIENTELAAWQRVQDVNLKSVYLCCRAALRHMVLAGKGSI
Rv2857c|M.tuberculosis_H37Rv AGISPP-EDNLIENTELAAWQRVQDVNLKSVYLCCRAALRHMVLAGKGSI
MSMEG_2598|M.smegmatis_MC2_155 AGISPP-EDDLIENTDLPAWQRVQDINLKSVYLSCRAALRHMVPAGKGSI
Mflv_4062|M.gilvum_PYR-GCK AGISPP-EDDLIENTGIDAWDRVQDINLKSVFFCCKAALRHMVPAQKGSI
Mvan_2281|M.vanbaalenii_PYR-1 AGISPP-EDDLIENTGVDAWDRVQDINLKSVFFCCKAALRHMVPAQKGSI
MMAR_4765|M.marinum_M AGVSGV-MHRRFLDDDLADFHRVMAVNVLGVMAGTRDAARHMAAHGGGSI
MUL_0336|M.ulcerans_Agy99 AGVSGA-MHRRFLDDDLADFHRVMAVNVLGVMAGTRDAARHMAAHGGGSI
TH_3693|M.thermoresistible__bu AGITRD-ATMRKMTEQ--QFDEVIAVHLKGTWNGLRAAAAIMREQKRGAI
MLBr_02565|M.leprae_Br4923 AGITRD---KLLANMDDCRWDAVLEVNLLAPQRLTEGLVGNGSISKGGRV
*.: :. . : . . * :
MAV_1832|M.avium_104 VNTASGAALKAAAG-MHAYVATKHGIVGLTRNAGLDYARNGIRVNAVAPG
MAB_1219|M.abscessus_ATCC_1997 VNQSSTAAWLYSG----FYGLAKVGINGLTQQLSRELGGQKIRINAIAPG
Mb2882c|M.bovis_AF2122/97 VNTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFARQGIRVNALCPG
Rv2857c|M.tuberculosis_H37Rv VNTASFVAVMGSATSQISYTASKGGVLAMSRELGVQFARQGIRVNALCPG
MSMEG_2598|M.smegmatis_MC2_155 INTASFVAVMGSATSQISYTASKGGVLAMSRELGVQYARQGIRVNALCPG
Mflv_4062|M.gilvum_PYR-GCK VNTASFVAVNGSATSQISYTASKGGVLAMSRELGIQYARQGIRVNALCPG
Mvan_2281|M.vanbaalenii_PYR-1 INTASFVAVMGSATSQISYTASKGGVLAMSRELGVQYARQGIRVNALCPG
MMAR_4765|M.marinum_M VNLTSIGGIQAGGG-VMTYRASKAAVIQFTKSAAIELAHYEIRVNAIAPG
MUL_0336|M.ulcerans_Agy99 VNLTSIGGIQAGGG-VMTYRASKGAVIQFTKSAAIELAHYEIRVNAIAPG
TH_3693|M.thermoresistible__bu VNMSSISGKVGMVG-QTNYSAAKAGIVGMTKAASKELAYLGVRVNAIQPG
MLBr_02565|M.leprae_Br4923 IVLSSMAGIAGNRG-QTNYATAKAGVIGLTQALAPGLAEKGITINAVAPG
: :* . * :* .: ::: . . : :**: **
MAV_1832|M.avium_104 SIKTPQMA-SFPQ---------EMQ--DEWERIIPMGRMGRPEEVADVVA
MAB_1219|M.abscessus_ATCC_1997 PTDTEATRGTVPE---------QFR--SELVKNIPLSRMGTVDDMVGMCL
Mb2882c|M.bovis_AF2122/97 PVNTPLLQELFAK---------NPERAARRMVHVPLGRFAEPDEIAAAVA
Rv2857c|M.tuberculosis_H37Rv PVNTPLLQELFAK---------NPERAARRMVHVPLGRFAEPDEIAAAVA
MSMEG_2598|M.smegmatis_MC2_155 PVNTPLLQELFAK---------DPERAARRLVHIPLGRFAEPEELAAAVA
Mflv_4062|M.gilvum_PYR-GCK PVNTPLLQELFAK---------DPERAARRLVHVPMGRFAEPEELAAAVA
Mvan_2281|M.vanbaalenii_PYR-1 PVNTPLLQELFAK---------DPERAARRLVHVPVGRFAEPEELAAAVA
MMAR_4765|M.marinum_M NIPTPLLASSAAGMDQEQVERFTAQIRQTMREDRPLKREGTPEDIAEAAL
MUL_0336|M.ulcerans_Agy99 NIPTPLLASSAAGLDQEQVERFTAQIRQTMREDRPLKREGTPEDIAEAAL
TH_3693|M.thermoresistible__bu -----LIRSAMT-------EAMPQRIWDEKLAEIPMGRAGEPEEVAKVAL
MLBr_02565|M.leprae_Br4923 FIETQMTAAIPLA----------TREVGRRLNSLLQG--GLPVDVAETIA
. . ::.
MAV_1832|M.avium_104 FLLSDAASFITATTVEIDGAYLQSSK-----
MAB_1219|M.abscessus_ATCC_1997 FLLSDKASWITGQVFNVDGGQIIRS------
Mb2882c|M.bovis_AF2122/97 FLASDDASFITASTFLVDGGISSAYVTPL--
Rv2857c|M.tuberculosis_H37Rv FLASDDASFITASTFLVDGGISSAYVTPL--
MSMEG_2598|M.smegmatis_MC2_155 FLASDDASFITGSTFLVDGGISSAYVTPL--
Mflv_4062|M.gilvum_PYR-GCK FLASDDASFITGSSFLVDGGITGHYVTPL--
Mvan_2281|M.vanbaalenii_PYR-1 FLASDDASFITASSFLVDGGISGHYVTPL--
MMAR_4765|M.marinum_M YFAGERSRYVTGTVLPVDGGTVAGKVIRRSR
MUL_0336|M.ulcerans_Agy99 YFAGERSRYVTGTVLPVDGGTVAGKVIRRSR
TH_3693|M.thermoresistible__bu FLASDLSSYMTGTVLEVTGGRHL--------
MLBr_02565|M.leprae_Br4923 YFAIPASNAVTGNVIRVCGQAMLGA------
:: : :*. . : *