For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
STSTQGTNPEDEAWPMIEEAVFRLFDELAAKGEHTVIGPRLNELGWAEIESEYPVEACALLFRAQGRSLA LTDALDRIMLAELASLFDEPVDAIVLPELTHGHTPSSTEDQVKGVVLGPLRGRVAVPVSRAMGTVSIAVV DADRLRGERMDTFDPSVVWSCVEGSTDGPLLDASTEWGRAIAAAHRSLATEVIALADQALRIAVDHISVR VQFGSPIGSLQSPRHALADAAARLEGARAVLSESWRYGGLLSAQTAKVAAGRAHRAVSDAVLQVCGAIGL TAEHDLHRYVSRGFQIDALCGSYLQLESLLAERLFDIYAPGRALPAIVNWD*
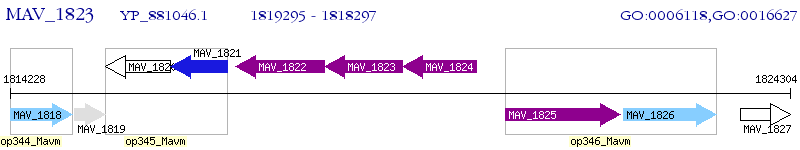
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1823 | - | - | 100% (332) | putative acyl-CoA dehydrogenase |
| M. avium 104 | MAV_2562 | - | e-123 | 68.40% (326) | putative acyl-CoA dehydrogenase |
| M. avium 104 | MAV_1631 | - | 3e-17 | 30.10% (299) | putative acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3594 | fadE33 | 1e-17 | 30.18% (275) | acyl-CoA dehydrogenase FADE33 |
| M. gilvum PYR-GCK | Mflv_2434 | - | 1e-16 | 27.36% (296) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3564 | fadE33 | 1e-17 | 30.18% (275) | acyl-CoA dehydrogenase FADE33 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 3e-06 | 27.93% (111) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0593c | - | 1e-13 | 36.97% (119) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_5053 | fadE33 | 9e-18 | 32.97% (279) | acyl-CoA dehydrogenase FadE33 |
| M. smegmatis MC2 155 | MSMEG_4845 | - | 1e-20 | 29.83% (295) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_4138 | - | 1e-144 | 78.37% (319) | PUTATIVE putative acyl-CoA dehydrogenase |
| M. ulcerans Agy99 | MUL_4129 | fadE33 | 3e-17 | 32.62% (279) | acyl-CoA dehydrogenase FadE33 |
| M. vanbaalenii PYR-1 | Mvan_4218 | - | 3e-19 | 29.73% (296) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5053|M.marinum_M -------MT----EEREMLRETVAALVNKHASPAAVRSAMESERGYDETL
MUL_4129|M.ulcerans_Agy99 -------MT----EEREMLRETVAALVNKHASPAAVRSAMESERGYDETL
Mb3594|M.bovis_AF2122/97 -------MTPP--EERQMLRETVASLVAKHAGPAAVRAAMASDRGYDESL
Rv3564|M.tuberculosis_H37Rv -------MTPP--EERQMLRETVASLVAKHAGPAAVRAAMASDRGYDESL
MAB_0593c|M.abscessus_ATCC_199 -------MSPSGNTERQMLRDTVRSLVTKRADSAAVRTAFESERGYDENL
MAV_1823|M.avium_104 ----MSTSTQGTNPEDEAWPMIEEAVFRLFDELAAKGEHTVIGPRLNELG
TH_4138|M.thermoresistible__bu ----MTAS-----QVDESWPMVEEAVFRLFDEIAAKQDHVAIDPLLSELG
Mflv_2434|M.gilvum_PYR-GCK ---MTVAIKDDAAMDEASLGMLEASLRKTMSSASGP----KLDAALTELG
Mvan_4218|M.vanbaalenii_PYR-1 ---------MAPGMDEASLDMLEASLRKTMLSSSGP----ELDAALTEMG
MSMEG_4845|M.smegmatis_MC2_155 -------------MDAESLGLLEETLRKTMLSASGA----KLDAALAELG
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRARFPEE
:: .
MMAR_5053|M.marinum_M WRLLCEQVGAAALVIPEELGGAGGELADAAIVLQELGRALVPSPLLGTT-
MUL_4129|M.ulcerans_Agy99 WRLLCEQVGAAALVIPEELGGAGGELADAAIVLQELGRALVPSPLLGTT-
Mb3594|M.bovis_AF2122/97 WRLLCEQVGAAALVIPEELGGAGGELADAAIVVQELGRALVPSPLLGTT-
Rv3564|M.tuberculosis_H37Rv WRLLCEQVGAAALVIPEELGGAGGELADAAIVVQELGRALVPSPLLGTT-
MAB_0593c|M.abscessus_ATCC_199 WAVLCEQVGAAALLIPEESGGAGGELADAAVVVEELGRGLVPSPLMGTL-
MAV_1823|M.avium_104 WAEIESEYPVEACALLFRAQGRSLALTDALDRIMLAELASLFDEPVDAI-
TH_4138|M.thermoresistible__bu WADIESEYPVEACALLFRAQGRSLALTDALDRVMIAEMASLVEEPVDCV-
Mflv_2434|M.gilvum_PYR-GCK WADMLTEAPELAIPLYFRLLGETGSHASILVDVVLHATG-----------
Mvan_4218|M.vanbaalenii_PYR-1 WADMLSEVPELAVPLVFRLLGETGSHASVLVDVVLHATG-----------
MSMEG_4845|M.smegmatis_MC2_155 WAEMLSDMPEVAMPLVFRLLGETGSHASVLNDVLLETIG-----------
MLBr_00737|M.leprae_Br4923 ALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIPAVN
* : . * . :
MMAR_5053|M.marinum_M ---LAELALLAAAEPDAKTLAGLAEGSSIGALVLDPDYVLNGDTADVVVA
MUL_4129|M.ulcerans_Agy99 ---LAELALLAAAEPDAKTLAGLAEGSSIGALVLDPDYVLNGDTADVVVA
Mb3594|M.bovis_AF2122/97 ---LAELALLAAAKPDAQALTELAQGSAIGALVLDPDYVVNGDIADIVVA
Rv3564|M.tuberculosis_H37Rv ---LAELALLAAAKPDAQALTELAQGSAIGALVLDPDYVVNGDIADIVVA
MAB_0593c|M.abscessus_ATCC_199 ---WAELALLEAGY-EGELLEALAAGESIGAVAFDPGYVVNGDIADVVLT
MAV_1823|M.avium_104 ---VLPELTHGHTPSSTEDQVKGVVLGPLRGRVAVPVSRAMGTVSIAVVD
TH_4138|M.thermoresistible__bu ---VLPALAQASVPSSNEGQVKGIVLGPLRGRVLVPVSGAMGTVSATVVD
Mflv_2434|M.gilvum_PYR-GCK --------------------------NAIGDTVELPLPYAGN--AWVVWD
Mvan_4218|M.vanbaalenii_PYR-1 --------------------------NTIGDTVELPLPYAGN--GWVVWD
MSMEG_4845|M.smegmatis_MC2_155 --------------------------GLPGGTP--PMPYAGG--GWVVWE
MLBr_00737|M.leprae_Br4923 KLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMRTRA
. . .
MMAR_5053|M.marinum_M STEG----------------------------QLSRWTRFSAHRVATMDP
MUL_4129|M.ulcerans_Agy99 STEG----------------------------QLSRWTRFSAHRVATMDP
Mb3594|M.bovis_AF2122/97 ATSG----------------------------QLTRWTRFSAQPVATMDP
Rv3564|M.tuberculosis_H37Rv ATSG----------------------------QLTRWTRFSAQPVATMDP
MAB_0593c|M.abscessus_ATCC_199 VQGE----------------------------TVCRAGQVTALALSTLDP
MAV_1823|M.avium_104 ADRL----------------------------RGERMDTFDPSVVWSCVE
TH_4138|M.thermoresistible__bu ADQL----------------------------RATRLNTFDSSVVWTRVE
Mflv_2434|M.gilvum_PYR-GCK RGGA----------------------------SEATGEDRGPESLDPTLG
Mvan_4218|M.vanbaalenii_PYR-1 RGGA----------------------------SEATGDDRSPESLDPTLG
MSMEG_4845|M.smegmatis_MC2_155 RRDS----------------------------TDS------ADSANPTLG
MLBr_00737|M.leprae_Br4923 KADGDDWILNGFKCWITNGGKSTWYTVMAVTDPDKGANGISAFIVHKDDE
.
MMAR_5053|M.marinum_M TRRLAQLRA--------------------------EETAFLGEDPGLADA
MUL_4129|M.ulcerans_Agy99 TRRLAQLRA--------------------------EETAFLGENPGLADA
Mb3594|M.bovis_AF2122/97 TRRLARLQS--------------------------EETEPLCPDPGIADT
Rv3564|M.tuberculosis_H37Rv TRRLARLQS--------------------------EETEPLCPDPGIADT
MAB_0593c|M.abscessus_ATCC_199 TRRLASIAV--------------------------GAAEPVGTAPGLIDK
MAV_1823|M.avium_104 GSTDGPLLD--------------------------ASTEWGRA----IAA
TH_4138|M.thermoresistible__bu GATDGELVD--------------------------ASTEWARA----VAA
Mflv_2434|M.gilvum_PYR-GCK G---LPLRR--------------------------EEEGYPIR----VAE
Mvan_4218|M.vanbaalenii_PYR-1 G---LPLRR--------------------------EEEGYPIR----LAE
MSMEG_4845|M.smegmatis_MC2_155 G---LPLRK--------------------------VPDGQLLR----ISE
MLBr_00737|M.leprae_Br4923 GFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGTGFKTALATLDH
MMAR_5053|M.marinum_M AAILLAAEQIGAAERCLELTVEYAKSRVQFGRPIGSFQALKHRMADLYVE
MUL_4129|M.ulcerans_Agy99 AGILLAAEQIGAAERCLELTVEYAKSRVQFGRPIGSFQALKHRMADLYVE
Mb3594|M.bovis_AF2122/97 AAILLAAEQIGAAERCLQLTVEYAKSRVQFGRPIGSFQALKHRMADLYVT
Rv3564|M.tuberculosis_H37Rv AAILLAAEQIGAAERCLQLTVEYAKSRVQFGRPIGSFQALKHRMADLYVT
MAB_0593c|M.abscessus_ATCC_199 AGLLLAAEQVGAAARALDLTVEYTKTRVQFGRAIGSFQALKHRMADLYVL
MAV_1823|M.avium_104 AHRSLATEVIALADQALRIAVDHISVRVQFGSPIGSLQSPRHALADAAAR
TH_4138|M.thermoresistible__bu AHRALATELIALADRALQIAVEHVSVRVQFGSPIGSLQSPRHLLAAAAAK
Mflv_2434|M.gilvum_PYR-GCK ARVAVGWWLVGSARAMLALARQHALDRVQFGKPIAGFQAVRHRLAETLVA
Mvan_4218|M.vanbaalenii_PYR-1 ARVAVGWWLVGSARAMLSLARQHALDRVQFGKPIASFQAVRHRLAETLVA
MSMEG_4845|M.smegmatis_MC2_155 ARRALGWWLVGSARAMLALARQHAMDRVQFGAPIATFQAVRHRLAETLVA
MLBr_00737|M.leprae_Br4923 TRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQFMLADMAMK
: :. :. * * : : * *** .*. :*: :. :*
MMAR_5053|M.marinum_M VAAARAVVTEACTAPTP------TSAATARLAACEALCAVAAEGIQLHGG
MUL_4129|M.ulcerans_Agy99 VAAARAVVTEACTAPTP------TSAATARLAACEALCAVAAEGIQLHGG
Mb3594|M.bovis_AF2122/97 IAAARAVVADACHAPTP------TNAATARLAASEALSTAAAEGIQLHGG
Rv3564|M.tuberculosis_H37Rv IAAARAVVADACHAPTP------TNAATARLAASEALSTAAAEGIQLHGG
MAB_0593c|M.abscessus_ATCC_199 VESARATVYQAIHDLTP------EGATVARIKASEALSAVSGDSIQLHGG
MAV_1823|M.avium_104 LEGARAVLSESWRYGGL------LSAQTAKVAAGRAHRAVSDAVLQVCGA
TH_4138|M.thermoresistible__bu LEGARALLDDSWHYGGV------LSAQVAKVSAGRAHRAVSDAVLQVCGA
Mflv_2434|M.gilvum_PYR-GCK IEGAEATLSLPSADNPD------LSSLLAKAAAGKAALTAAKNCQQVLGG
Mvan_4218|M.vanbaalenii_PYR-1 IEGAEATLSLPGADNPD------LTALLAKAAAGKAALTAAMHCQQVLGG
MSMEG_4845|M.smegmatis_MC2_155 IEGAEATLSQPNADNVD------LTSLLAKAAAGKAALTAARHCQQVLGG
MLBr_00737|M.leprae_Br4923 VEAARLIVYAAAARAERGEPDLGFISAASKCFASDIAMEVTTDAVQLFGG
: .*. : . : :: * .: *: *.
MMAR_5053|M.marinum_M IAITWEHDMHLYFKRAHGSTHLLASPPELLRRLESEALGAGPSIP-----
MUL_4129|M.ulcerans_Agy99 IAITWEHDMHLYFKRAHGSTHLLASPPELLRRLESEALGAGPSIP-----
Mb3594|M.bovis_AF2122/97 IAITWEHDMHLYFKRAHGSAQLLESPREVLRRLESEVWES-P--------
Rv3564|M.tuberculosis_H37Rv IAITWEHDMHLYFKRAHGSAQLLESPREVLRRLESEVWES-P--------
MAB_0593c|M.abscessus_ATCC_199 IAITWEHDAHLYFKRAHGSALLLGTPRQLLPNLETAAGL-----------
MAV_1823|M.avium_104 IGLTAEHDLHRYVSRGFQIDALCGSYLQLESLLAERLFDIYAPGRALPAI
TH_4138|M.thermoresistible__bu VGLTAEHDLHRYVRRGMQIDSLCGSYRQLESLLADRLSAIYAPGRALPAI
Mflv_2434|M.gilvum_PYR-GCK IGFTEEHGLQHHVKRTLVLDGLLGSSRELTRRAGAGLRARGSVPRLAQL-
Mvan_4218|M.vanbaalenii_PYR-1 IGFTEEHHLHRHVKRALVLDGLLGSSRELTRRAGAGLRARGSAPRLAQL-
MSMEG_4845|M.smegmatis_MC2_155 IGFTAEHELHTHVERVLVLDGLLGNARELTRKAGSGMRARGTAPRMAQV-
MLBr_00737|M.leprae_Br4923 AGYTSDFPVERFMRDAKITQIYEGTN-QIQRVVMSRALLR----------
. * :. . .. . ::
MMAR_5053|M.marinum_M ----
MUL_4129|M.ulcerans_Agy99 ----
Mb3594|M.bovis_AF2122/97 ----
Rv3564|M.tuberculosis_H37Rv ----
MAB_0593c|M.abscessus_ATCC_199 ----
MAV_1823|M.avium_104 VNWD
TH_4138|M.thermoresistible__bu INVN
Mflv_2434|M.gilvum_PYR-GCK ----
Mvan_4218|M.vanbaalenii_PYR-1 ----
MSMEG_4845|M.smegmatis_MC2_155 ----
MLBr_00737|M.leprae_Br4923 ----