For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KTDLPQDISDFATVAAKRFTRLGGPPAALRAETDDTVREAARTALAEVGAFDLDVRTAPDDLLAAAVLCQ AAGAAALPYPLVEELLAIDGARLTLVNPAAPRIDHGDLAGDWIAADIDGNRYRPRPGSRTGAKLGPFLVP ATLSAPEGTVPGADVNLHLVLGAWRILGAVQQSLRIVTDHVQARIQFGKPLADFQAVRFAVADAAVMVRG LHELAKYTICRPESLPAQVHSADALVLRLKAADTARQVMRTSHQLLGALGFCDESDVSVLDRHTQPLIRL PLSTEELALRLIPSVADGAFETLFSEPVSA*
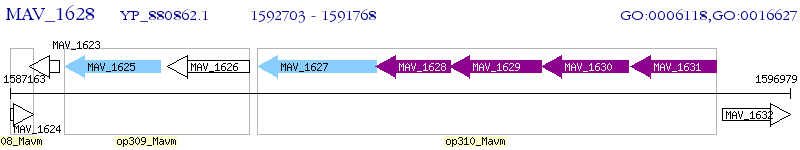
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_1628 | - | - | 100% (311) | putative acyl-CoA dehydrogenase |
| M. avium 104 | MAV_1336 | - | 1e-10 | 30.95% (210) | acyl-CoA dehydrogenase domain-containing protein |
| M. avium 104 | MAV_0597 | - | 8e-10 | 27.44% (317) | putative acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3594 | fadE33 | 1e-09 | 24.29% (317) | acyl-CoA dehydrogenase FADE33 |
| M. gilvum PYR-GCK | Mflv_1793 | - | 2e-42 | 39.44% (284) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3564 | fadE33 | 1e-09 | 24.29% (317) | acyl-CoA dehydrogenase FADE33 |
| M. leprae Br4923 | MLBr_00661 | fadE24 | 6e-05 | 34.65% (101) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0614 | - | 2e-09 | 40.62% (96) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_3982 | - | 1e-136 | 78.14% (311) | acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5621 | - | 8e-40 | 39.86% (286) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_0680 | - | 2e-39 | 38.28% (303) | putative acyl-CoA dehydrogenase |
| M. ulcerans Agy99 | MUL_3843 | - | 1e-134 | 76.85% (311) | acyl-CoA dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4957 | - | 1e-40 | 39.30% (285) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3594|M.bovis_AF2122/97 --------------------------------------------------
Rv3564|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3982|M.marinum_M --------------------------------------------------
MUL_3843|M.ulcerans_Agy99 --------------------------------------------------
MAV_1628|M.avium_104 --------------------------------------------------
Mflv_1793|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4957|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5621|M.smegmatis_MC2_155 --------------------------------------------------
TH_0680|M.thermoresistible__bu --------------------------------------------------
MAB_0614|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00661|M.leprae_Br4923 MTDTAPSAKPSSVRLKRGGLRSGVGMQPHKRTGFDVAIAMLTPLVGQDFL
Mb3594|M.bovis_AF2122/97 --------------------------------------------------
Rv3564|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3982|M.marinum_M --------------------------------------------------
MUL_3843|M.ulcerans_Agy99 --------------------------------------------------
MAV_1628|M.avium_104 --------------------------------------------------
Mflv_1793|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4957|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5621|M.smegmatis_MC2_155 --------------------------------------------------
TH_0680|M.thermoresistible__bu -----------------------------------------------MDG
MAB_0614|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_00661|M.leprae_Br4923 DKYHLRYPLNRALRYGVKATFSTAAATSRQFKRVQNLRGGATRLQPSGRN
Mb3594|M.bovis_AF2122/97 ---MTPPEERQMLRETVASLVAK---------------------------
Rv3564|M.tuberculosis_H37Rv ---MTPPEERQMLRETVASLVAK---------------------------
MMAR_3982|M.marinum_M -MNTELPQDIIEFARVAAKRLAR---------------------------
MUL_3843|M.ulcerans_Agy99 -MNTELPQDVIDFARVAAKRLAR---------------------------
MAV_1628|M.avium_104 -MKTDLPQDISDFATVAAKRFTR---------------------------
Mflv_1793|M.gilvum_PYR-GCK -MILDLSDDALEFGRQALHAFEN---------------------------
Mvan_4957|M.vanbaalenii_PYR-1 -MILDLSDDALEYGLQALHAFEN---------------------------
MSMEG_5621|M.smegmatis_MC2_155 -MILDLSDDAEEFGHQALRAFES---------------------------
TH_0680|M.thermoresistible__bu PMILDLSDDALEYGRQALRAFEA---------------------------
MAB_0614|M.abscessus_ATCC_1997 -MDFAFAEEQQAVSDVVEAVLADREFVGVVPEIGYDEAAWRALAANGVLS
MLBr_00661|M.leprae_Br4923 YFDLTPDDNQKLIVETIDEFSAEILR------------------------
::
Mb3594|M.bovis_AF2122/97 ------HAGP-------AAVRAAMASDRGYDESLWRLLCEQVGAAALVIP
Rv3564|M.tuberculosis_H37Rv ------HAGP-------AAVRAAMASDRGYDESLWRLLCEQVGAAALVIP
MMAR_3982|M.marinum_M ------VGGP-------PAALRAETDD-GIRRAAR-AALDDLGAFELDVR
MUL_3843|M.ulcerans_Agy99 ------VGGP-------PAALRAETDD-GIRRAAC-AALDDLGAFELDVR
MAV_1628|M.avium_104 ------LGGP-------PAALRAETDD-TVREAAR-TALAEVGAFDLDVR
Mflv_1793|M.gilvum_PYR-GCK ------AGGD-------RLAEQAEADP-AARQTLVAPVLDGLGAWDLDPR
Mvan_4957|M.vanbaalenii_PYR-1 ------AGGD-------RLVEQAEADP-TARHGLVRPVLDGLGAWDLDPR
MSMEG_5621|M.smegmatis_MC2_155 ------AGGD-------ELLQQAEAKP-DARQGLVGPVLDGLGAWDLDPR
TH_0680|M.thermoresistible__bu ------AGGD-------ELVLQADAKP-ENREALVGPVLTGLDAWDLDPR
MAB_0614|M.abscessus_ATCC_1997 LALPERLGGDGLGLAEVSVALRVLGKHGALTPALATLGFGVVPLVELASG
MLBr_00661|M.leprae_Br4923 -----PAAHDADEAAAYPPDLIAKAAKLGITTINIPEYFDGIAERRSSVT
. . :
Mb3594|M.bovis_AF2122/97 ----------------------------------EELGGAGGELADAAIV
Rv3564|M.tuberculosis_H37Rv ----------------------------------EELGGAGGELADAAIV
MMAR_3982|M.marinum_M ----------------------------------SGPD----DLLAAAVL
MUL_3843|M.ulcerans_Agy99 ----------------------------------SGPD----DLLAAAVL
MAV_1628|M.avium_104 ----------------------------------TAPD----DLLAAAVL
Mflv_1793|M.gilvum_PYR-GCK ----------------------------------TDPD----GLEAAAAL
Mvan_4957|M.vanbaalenii_PYR-1 ----------------------------------TDAD----GLEAAAAL
MSMEG_5621|M.smegmatis_MC2_155 ----------------------------------SDAD----ALEAAAAL
TH_0680|M.thermoresistible__bu ----------------------------------TDLD----GAEAAAAL
MAB_0614|M.abscessus_ATCC_1997 ----------------------------------EQQDRYLAGVGDGAIL
MLBr_00661|M.leprae_Br4923 NVLVAEALAYGDMGLTLPILAPGGVAATLTYWGSADQQATYLKEFTGENV
. :
Mb3594|M.bovis_AF2122/97 VQELGRALVPSPLLGTTLAELALLAAAKPDAQALTELAQGSAIGALVLDP
Rv3564|M.tuberculosis_H37Rv VQELGRALVPSPLLGTTLAELALLAAAKPDAQALTELAQGSAIGALVLDP
MMAR_3982|M.marinum_M CRAAGALAIPYPLVEE------LLS---VDGARLALVNP---------DL
MUL_3843|M.ulcerans_Agy99 CRAAGTVALPYPLVEE------LLS---IDGARLALVNP---------DL
MAV_1628|M.avium_104 CQAAGAAALPYPLVEE------LLA---IDGARLTLVNP---------AA
Mflv_1793|M.gilvum_PYR-GCK CRSAGYWAVPYPLAER------LAAPTDLDVDGLLVIPA---------TR
Mvan_4957|M.vanbaalenii_PYR-1 CRSAGYWALPYPLAER------LAKPADLDVDGLVVVGG---------PR
MSMEG_5621|M.smegmatis_MC2_155 CRSAGYWALPYPVAER------LARPTDLDVDGLIVVAD---------GT
TH_0680|M.thermoresistible__bu CRSAGYWALPYPVAER------LSRPTDLDVDGLIVVDA---------AA
MAB_0614|M.abscessus_ATCC_1997 TAALDEPGRAVPSAPA--------VSAVRDGSSLVLNGRKIGVLHAAEAH
MLBr_00661|M.leprae_Br4923 PQACVAIAEPQPLFDPTR----LKTIAVRTPSGYRLDGVKSLVPAAASAE
. *
Mb3594|M.bovis_AF2122/97 DYVVNGDIADI----VVAATSGQLTRWTRFSAQPVATMDPTRRLARLQSE
Rv3564|M.tuberculosis_H37Rv DYVVNGDIADI----VVAATSGQLTRWTRFSAQPVATMDPTRRLARLQSE
MMAR_3982|M.marinum_M PRIDHGDLPGS----WIAADLDGARYRPQPAARTAAKLGPFLVPATLGAP
MUL_3843|M.ulcerans_Agy99 PRIDHGDLPGS----WIAADLDGARYRPQPAARTAAKLGPFLVPATLGAP
MAV_1628|M.avium_104 PRIDHGDLAGD----WIAADIDGNRYRPRPGSRTGAKLGPFLVPATLSAP
Mflv_1793|M.gilvum_PYR-GCK PSAPVAGLDLR----WAAVDLDGRRS----TVTGHGEPGPAFVAELELSP
Mvan_4957|M.vanbaalenii_PYR-1 PAAALGGLDLR----WAAVDLDGRRA----LVTGQGVTGPGFVAELELSP
MSMEG_5621|M.smegmatis_MC2_155 PEAALAGLDLR----WAAVTLDGTRS----TVTRLGATGPAFTTALDLVE
TH_0680|M.thermoresistible__bu PLGGVAGVRLR----WATVDLTGRRA----DVTQVVGAGPGFTARLETTP
MAB_0614|M.abscessus_ATCC_1997 WILVTTDNGVV----VVAPNTPGVTITRTPTASKAAEYAVTFSDVSVPAA
MLBr_00661|M.leprae_Br4923 LFIIGAQLNGKPALFIVESSTKGLTVKADPSMGIRAAALGQVELCGVSAP
Mb3594|M.bovis_AF2122/97 ETEPLCPDPG-------IADTAAILLAAEQIGAAERCLQLTVEYAKSRVQ
Rv3564|M.tuberculosis_H37Rv ETEPLCPDPG-------IADTAAILLAAEQIGAAERCLQLTVEYAKSRVQ
MMAR_3982|M.marinum_M EGTVAASD----------IDLHLVLGSWRILGAVQQSLDIVTAHVRTRIQ
MUL_3843|M.ulcerans_Agy99 EGTVAASD----------IDLHLVLGSWRILGAVQQTLDIVTAHVRTRIQ
MAV_1628|M.avium_104 EGTVPGAD----------VNLHLVLGAWRILGAVQQSLRIVTDHVQARIQ
Mflv_1793|M.gilvum_PYR-GCK VDDAGGPE----------LALALVLPSWTLLGMLDRALALTVSHVGLRKQ
Mvan_4957|M.vanbaalenii_PYR-1 VDSDGSRD----------VALALVLPCWTLLGILDRAVALTVAHVSLRKQ
MSMEG_5621|M.smegmatis_MC2_155 IDAQGAGD----------VALGLVLGSWTLLGLLDRAVDLTVAHVSLRKQ
TH_0680|M.thermoresistible__bu LDEAGGDD----------IALALVLSGWTLLGMLDRAIGLTVEHVRLREQ
MAB_0614|M.abscessus_ATCC_1997 DQLSGSVQR--------LNELALAAIGSYADGLVSGALRLTADHVSNRVQ
MLBr_00661|M.leprae_Br4923 LDARLGEDDATDNDYSEAIALARLGWAALAVGTSHAVLDYVMPYVKEREA
: * : . :. *
Mb3594|M.bovis_AF2122/97 FGRPIGSFQALKHRMADLYVTIAAARAVVADACHAPT-------PTNAAT
Rv3564|M.tuberculosis_H37Rv FGRPIGSFQALKHRMADLYVTIAAARAVVADACHAPT-------PTNAAT
MMAR_3982|M.marinum_M FGKALADFQAVRFCVADAAVAVRGLAELAKYTISRTESYAARVRSADALL
MUL_3843|M.ulcerans_Agy99 LGKALADFQTVRFCVANAAVAVRGLAELAKYTISRTESYAARVRSADALL
MAV_1628|M.avium_104 FGKPLADFQAVRFAVADAAVMVRGLHELAKYTICRPESLPAQVHSADALV
Mflv_1793|M.gilvum_PYR-GCK FGQTLSSFQGVQFQLTDAEVERSGVDILAKHALWSIA-TDQPQAVDDALA
Mvan_4957|M.vanbaalenii_PYR-1 FGQTLSSFQGVQFQLTDAEVERSGVDILAKHALWSIA-TDQSHVVDDALA
MSMEG_5621|M.smegmatis_MC2_155 FGQTLSSFQGVQFQLTDAEVERSGVDILAKHALWGLTRHDTTEVVDDALA
TH_0680|M.thermoresistible__bu FGQPLARFQGVQFQLTDAEVERSGTDILAKHALWSVS-TGQPGALEDALS
MAB_0614|M.abscessus_ATCC_1997 FGKPLATFQAVSQQLADVYVVSRTLNLGVTSAVWRLC--EGLDATEDLDI
MLBr_00661|M.leprae_Br4923 FGEPIARRQAVAFMCANIAIELDGLRLITWRGASRAE--QGLSFAREAAL
:*..:. * : :: : . :
Mb3594|M.bovis_AF2122/97 ARLAASEALSTAAAEGIQLHGGIAITWEHDMHLYFKRAHGSAQLLESPRE
Rv3564|M.tuberculosis_H37Rv ARLAASEALSTAAAEGIQLHGGIAITWEHDMHLYFKRAHGSAQLLESPRE
MMAR_3982|M.marinum_M LRLKAADTARQVLRISHQLLGALGFCDESDISVLDRHTQPLIRLPLCAEA
MUL_3843|M.ulcerans_Agy99 LRLKAADTARQVLRISHQLLGALGFCDESDISVLDRHTQPLIRLPLCAEA
MAV_1628|M.avium_104 LRLKAADTARQVMRTSHQLLGALGFCDESDVSVLDRHTQPLIRLPLSTEE
Mflv_1793|M.gilvum_PYR-GCK LRLAALEAAEVVFRVCHQLHGAAGFCDETTLSWLSRYSQPLRRLPFGLSA
Mvan_4957|M.vanbaalenii_PYR-1 LRMAAIEAADVVFRVCHQLHGAVGFCDETTLSWLSRYSLPLRRLPFGLSS
MSMEG_5621|M.smegmatis_MC2_155 LRLAAIEAAEVVFRVCHQLHGAVGFCDETTLSWLSRHSQPLRRLPFGIST
TH_0680|M.thermoresistible__bu FRMAALEAAEVVFRVCHQLHGAVGFCDETTLSWLSRYSQPLRRLPLGVSA
MAB_0614|M.abscessus_ATCC_1997 VSFWMAAEAQRVMQICHHLHGGLGVDITYPMNRYYSTIKDLSRLVGGSSE
MLBr_00661|M.leprae_Br4923 AKRLGSDKAMQIGLDGVQLLGGHGFIKEHPVERWYRDLR-----AIGVAE
:* *. .. :
Mb3594|M.bovis_AF2122/97 VLRRLESEVWESP-----------
Rv3564|M.tuberculosis_H37Rv VLRRLESEVWESP-----------
MMAR_3982|M.marinum_M LAIRLIPDAADGSLETLFSAPVSA
MUL_3843|M.ulcerans_Agy99 LAIRLIPDAADGSLETLFSAPVSA
MAV_1628|M.avium_104 LALRLIPSVADGAFETLFSEPVSA
Mflv_1793|M.gilvum_PYR-GCK TRDHLTRLAGRRGLAGLYT-----
Mvan_4957|M.vanbaalenii_PYR-1 TRDHLTRVAGRRGLTGLFI-----
MSMEG_5621|M.smegmatis_MC2_155 TRDILTRRVGRRGLTGLFS-----
TH_0680|M.thermoresistible__bu TRAELSRRLGRRGLTGLYA-----
MAB_0614|M.abscessus_ATCC_1997 RLDVLAAAQG--------------
MLBr_00661|M.leprae_Br4923 GVVVI-------------------
: