For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ILDLSDDAEEFGHQALRAFESAGGDELLQQAEAKPDARQGLVGPVLDGLGAWDLDPRSDADALEAAAALC RSAGYWALPYPVAERLARPTDLDVDGLIVVADGTPEAALAGLDLRWAAVTLDGTRSTVTRLGATGPAFTT ALDLVEIDAQGAGDVALGLVLGSWTLLGLLDRAVDLTVAHVSLRKQFGQTLSSFQGVQFQLTDAEVERSG VDILAKHALWGLTRHDTTEVVDDALALRLAAIEAAEVVFRVCHQLHGAVGFCDETTLSWLSRHSQPLRRL PFGISTTRDILTRRVGRRGLTGLFS*
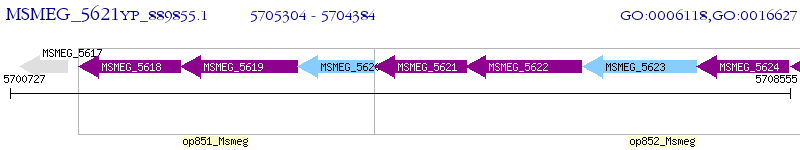
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5621 | - | - | 100% (306) | putative acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_5907 | - | 9e-12 | 32.57% (175) | acyl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6015 | - | 7e-11 | 26.67% (240) | putative acyl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3574c | fadE28 | 2e-11 | 30.00% (150) | acyl-CoA dehydrogenase FADE28 |
| M. gilvum PYR-GCK | Mflv_1793 | - | 1e-130 | 76.14% (306) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv3544c | fadE28 | 2e-11 | 30.00% (150) | acyl-CoA dehydrogenase FADE28 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 6e-06 | 24.63% (134) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0614 | - | 7e-12 | 30.54% (203) | acyl-CoA dehydrogenase FadE |
| M. marinum M | MMAR_3982 | - | 1e-43 | 40.00% (285) | acyl-CoA dehydrogenase |
| M. avium 104 | MAV_1628 | - | 6e-40 | 39.86% (286) | putative acyl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_0680 | - | 1e-122 | 70.26% (306) | putative acyl-CoA dehydrogenase |
| M. ulcerans Agy99 | MUL_3843 | - | 5e-42 | 38.95% (285) | acyl-CoA dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_4957 | - | 1e-134 | 78.69% (305) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1793|M.gilvum_PYR-GCK -----------------------------MILDLSDDALEF---------
Mvan_4957|M.vanbaalenii_PYR-1 -----------------------------MILDLSDDALEY---------
MSMEG_5621|M.smegmatis_MC2_155 -----------------------------MILDLSDDAEEF---------
TH_0680|M.thermoresistible__bu -------------------------MDGPMILDLSDDALEY---------
MMAR_3982|M.marinum_M -----------------------------MNTELPQDIIEF---------
MUL_3843|M.ulcerans_Agy99 -----------------------------MNTELPQDVIDF---------
MAV_1628|M.avium_104 -----------------------------MKTDLPQDISDF---------
Mb3574c|M.bovis_AF2122/97 -------------------MDFDPTAEQQAVADVVTSVLER---------
Rv3544c|M.tuberculosis_H37Rv -------------------MDFDPTAEQQAVADVVTSVLER---------
MAB_0614|M.abscessus_ATCC_1997 -------------------MDFAFAEEQQAVSDVVEAVLADREFVGVVPE
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRAR----
::
Mflv_1793|M.gilvum_PYR-GCK --------------GRQALHAFENAGGD---RLAEQAEADPAARQ---TL
Mvan_4957|M.vanbaalenii_PYR-1 --------------GLQALHAFENAGGD---RLVEQAEADPTARH---GL
MSMEG_5621|M.smegmatis_MC2_155 --------------GHQALRAFESAGGD---ELLQQAEAKPDARQ---GL
TH_0680|M.thermoresistible__bu --------------GRQALRAFEAAGGD---ELVLQADAKPENRE---AL
MMAR_3982|M.marinum_M --------------ARVAAKRLARVGGP---PAALRAETDDGIRR---AA
MUL_3843|M.ulcerans_Agy99 --------------ARVAAKRLARVGGP---PAALRAETDDGIRR---AA
MAV_1628|M.avium_104 --------------ATVAAKRFTRLGGP---PAALRAETDDTVRE---AA
Mb3574c|M.bovis_AF2122/97 ----DISWEALVCGGVTALPVPERLGGDGVGLFEVGALLTEVGRH---GA
Rv3544c|M.tuberculosis_H37Rv ----DISWEALVCGGVTALPVPERLGGDGVGLFEVGALLTEVGRH---GA
MAB_0614|M.abscessus_ATCC_1997 IGYDEAAWRALAANGVLSLALPERLGGDGLGLAEVSVALRVLGKH---GA
MLBr_00737|M.leprae_Br4923 --FPEEALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASAS
. : ** :
Mflv_1793|M.gilvum_PYR-GCK VAPVLDGLGA--------WDLDPRTDPD----GLEAAAALCRSAGYWAVP
Mvan_4957|M.vanbaalenii_PYR-1 VRPVLDGLGA--------WDLDPRTDAD----GLEAAAALCRSAGYWALP
MSMEG_5621|M.smegmatis_MC2_155 VGPVLDGLGA--------WDLDPRSDAD----ALEAAAALCRSAGYWALP
TH_0680|M.thermoresistible__bu VGPVLTGLDA--------WDLDPRTDLD----GAEAAAALCRSAGYWALP
MMAR_3982|M.marinum_M R-AALDDLGA--------FELDVRSGPD----DLLAAAVLCRAAGALAIP
MUL_3843|M.ulcerans_Agy99 C-AALDDLGA--------FELDVRSGPD----DLLAAAVLCRAAGTVALP
MAV_1628|M.avium_104 R-TALAEVGA--------FDLDVRTAPD----DLLAAAVLCQAAGAAALP
Mb3574c|M.bovis_AF2122/97 VTPALATLGLG---VVPLLELASAEQQDRFLAGVAKGGVLTAALNEPGAA
Rv3544c|M.tuberculosis_H37Rv VTPALATLGLG---VVPLLELASAEQQDRFLAGVAKGGVLTAALNEPGAA
MAB_0614|M.abscessus_ATCC_1997 LTPALATLGFG---VVPLVELASGEQQDRYLAGVGDGAILTAALDEPGRA
MLBr_00737|M.leprae_Br4923 LIPAVNKLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAA
..: :. :* . . * .
Mflv_1793|M.gilvum_PYR-GCK YPLAERLAAPTD-----LDVDGLLVIPATRPSAPVAGLDL---------R
Mvan_4957|M.vanbaalenii_PYR-1 YPLAERLAKPAD-----LDVDGLVVVGGPRPAAALGGLDL---------R
MSMEG_5621|M.smegmatis_MC2_155 YPVAERLARPTD-----LDVDGLIVVADGTPEAALAGLDL---------R
TH_0680|M.thermoresistible__bu YPVAERLSRPTD-----LDVDGLIVVDAAAPLGGVAGVRL---------R
MMAR_3982|M.marinum_M YPLVEELLS--------VDGARLALVNPDLPRIDHGDLPG---------S
MUL_3843|M.ulcerans_Agy99 YPLVEELLS--------IDGARLALVNPDLPRIDHGDLPG---------S
MAV_1628|M.avium_104 YPLVEELLA--------IDGARLTLVNPAAPRIDHGDLAG---------D
Mb3574c|M.bovis_AF2122/97 LPDRPATSFVGG----RLSGTKVGVGYAEQADWMLVTADN---------A
Rv3544c|M.tuberculosis_H37Rv LPDRPATSFVGG----RLSGTKVGVGYAEQADWMLVTADN---------A
MAB_0614|M.abscessus_ATCC_1997 VPSAPAVSAVRDGSSLVLNGRKIGVLHAAEAHWILVTTDN---------G
MLBr_00737|M.leprae_Br4923 SMRTRAKADGDD---WILNGFKCWITNGGKSTWYTVMAVTDPDKGANGIS
:. : .
Mflv_1793|M.gilvum_PYR-GCK WAAVDLDGRRS----TVTGH---GEPGPAFVAELELSPVDDAGGP-----
Mvan_4957|M.vanbaalenii_PYR-1 WAAVDLDGRRA----LVTGQ---GVTGPGFVAELELSPVDSDGSR-----
MSMEG_5621|M.smegmatis_MC2_155 WAAVTLDGTRS----TVTRL---GATGPAFTTALDLVEIDAQGAG-----
TH_0680|M.thermoresistible__bu WATVDLTGRRA----DVTQV---VGAGPGFTARLETTPLDEAGGD-----
MMAR_3982|M.marinum_M WIAADLDGARYRPQPAARTA---AKLGPFLVPATLGAPEGTVAAS-----
MUL_3843|M.ulcerans_Agy99 WIAADLDGARYRPQPAARTA---AKLGPFLVPATLGAPEGTVAAS-----
MAV_1628|M.avium_104 WIAADIDGNRYRPRPGSRTG---AKLGPFLVPATLSAPEGTVPGA-----
Mb3574c|M.bovis_AF2122/97 VVVVSPTADGVRMVRTPTSN---GSDEYVMTMDGVAVADCDILADV----
Rv3544c|M.tuberculosis_H37Rv VVVVSPTADGVRMVRTPTSN---GSDEYVMTMDGVAVADCDILADV----
MAB_0614|M.abscessus_ATCC_1997 VVVVAPNTPGVTITRTPTAS---KAAEYAVTFSDVSVPAADQLSG-----
MLBr_00737|M.leprae_Br4923 AFIVHKDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGTGF
. . .
Mflv_1793|M.gilvum_PYR-GCK -----ELALALVLPSWTLLGMLDRALALTVSHVGLRKQFGQTLSSFQGVQ
Mvan_4957|M.vanbaalenii_PYR-1 -----DVALALVLPCWTLLGILDRAVALTVAHVSLRKQFGQTLSSFQGVQ
MSMEG_5621|M.smegmatis_MC2_155 -----DVALGLVLGSWTLLGLLDRAVDLTVAHVSLRKQFGQTLSSFQGVQ
TH_0680|M.thermoresistible__bu -----DIALALVLSGWTLLGMLDRAIGLTVEHVRLREQFGQPLARFQGVQ
MMAR_3982|M.marinum_M -----DIDLHLVLGSWRILGAVQQSLDIVTAHVRTRIQFGKALADFQAVR
MUL_3843|M.ulcerans_Agy99 -----DIDLHLVLGSWRILGAVQQTLDIVTAHVRTRIQLGKALADFQTVR
MAV_1628|M.avium_104 -----DVNLHLVLGAWRILGAVQQSLRIVTDHVQARIQFGKPLADFQAVR
Mb3574c|M.bovis_AF2122/97 -AAHRVNQLALAVMGAYADGLVAGALRLTADYVANRKQFGKPLSTFQTVA
Rv3544c|M.tuberculosis_H37Rv -AAHRVNQLALAVMGAYADGLVAGALRLTADYVANRKQFGKPLSTFQTVA
MAB_0614|M.abscessus_ATCC_1997 -SVQRLNELALAAIGSYADGLVSGALRLTADHVSNRVQFGKPLATFQAVS
MLBr_00737|M.leprae_Br4923 KTALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQ
* :: . :. * *:*:.:: ** :
Mflv_1793|M.gilvum_PYR-GCK FQLTDAEVERSGVDILAKHALWSIA-TDQPQAVDDALALRLAALEAAEVV
Mvan_4957|M.vanbaalenii_PYR-1 FQLTDAEVERSGVDILAKHALWSIA-TDQSHVVDDALALRMAAIEAADVV
MSMEG_5621|M.smegmatis_MC2_155 FQLTDAEVERSGVDILAKHALWGLTRHDTTEVVDDALALRLAAIEAAEVV
TH_0680|M.thermoresistible__bu FQLTDAEVERSGTDILAKHALWSVS-TGQPGALEDALSFRMAALEAAEVV
MMAR_3982|M.marinum_M FCVADAAVAVRGLAELAKYTISRTESYAARVRSADALLLRLKAADTARQV
MUL_3843|M.ulcerans_Agy99 FCVANAAVAVRGLAELAKYTISRTESYAARVRSADALLLRLKAADTARQV
MAV_1628|M.avium_104 FAVADAAVMVRGLHELAKYTICRPESLPAQVHSADALVLRLKAADTARQV
Mb3574c|M.bovis_AF2122/97 AQLAEVYIASRTIDLVAKSVIWRLA--EDLDAGDDLGVLGYWVTSQAPPA
Rv3544c|M.tuberculosis_H37Rv AQLAEVYIASRTIDLVAKSVIWRLA--EDLDAGDDLGVLGYWVTSQAPPA
MAB_0614|M.abscessus_ATCC_1997 QQLADVYVVSRTLNLGVTSAVWRLC--EGLDATEDLDIVSFWMAAEAQRV
MLBr_00737|M.leprae_Br4923 FMLADMAMKVEAARLIVYAAAARAE-RGEPDLGFISAASKCFASDIAMEV
::: : . . * .
Mflv_1793|M.gilvum_PYR-GCK FRVCHQLHGAAGFCDETTLSWLSRYSQPLRRLPFGLSATRDHLTRLAGRR
Mvan_4957|M.vanbaalenii_PYR-1 FRVCHQLHGAVGFCDETTLSWLSRYSLPLRRLPFGLSSTRDHLTRVAGRR
MSMEG_5621|M.smegmatis_MC2_155 FRVCHQLHGAVGFCDETTLSWLSRHSQPLRRLPFGISTTRDILTRRVGRR
TH_0680|M.thermoresistible__bu FRVCHQLHGAVGFCDETTLSWLSRYSQPLRRLPLGVSATRAELSRRLGRR
MMAR_3982|M.marinum_M LRISHQLLGALGFCDESDISVLDRHTQPLIRLPLCAEALAIRLIPDAADG
MUL_3843|M.ulcerans_Agy99 LRISHQLLGALGFCDESDISVLDRHTQPLIRLPLCAEALAIRLIPDAADG
MAV_1628|M.avium_104 MRTSHQLLGALGFCDESDVSVLDRHTQPLIRLPLSTEELALRLIPSVADG
Mb3574c|M.bovis_AF2122/97 MQICHHLHGGMGMDVTYPMHRYYSTIKDLTRLLGGPSHRLELLGARCSLT
Rv3544c|M.tuberculosis_H37Rv MQICHHLHGGMGMDVTYPMHRYYSTIKDLTRLLGGPSHRLELLGARCSLT
MAB_0614|M.abscessus_ATCC_1997 MQICHHLHGGLGVDITYPMNRYYSTIKDLSRLVGGSSERLDVLAAAQG--
MLBr_00737|M.leprae_Br4923 TTDAVQLFGGAGYTSDFPVERFMRDAK-ITQIYEGTNQIQRVVMSRALLR
. :* *. * : : :: . :
Mflv_1793|M.gilvum_PYR-GCK GLAGLYT-----
Mvan_4957|M.vanbaalenii_PYR-1 GLTGLFI-----
MSMEG_5621|M.smegmatis_MC2_155 GLTGLFS-----
TH_0680|M.thermoresistible__bu GLTGLYA-----
MMAR_3982|M.marinum_M SLETLFSAPVSA
MUL_3843|M.ulcerans_Agy99 SLETLFSAPVSA
MAV_1628|M.avium_104 AFETLFSEPVSA
Mb3574c|M.bovis_AF2122/97 ------------
Rv3544c|M.tuberculosis_H37Rv ------------
MAB_0614|M.abscessus_ATCC_1997 ------------
MLBr_00737|M.leprae_Br4923 ------------