For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKRLEGKRILVTGAASGIGQATALRLLDEGAAVAASDVAVDGLERTRDRAAQAGTAERLVTVTMDVGRED AVVDGVRVAVEQLGGLDSLVNAAGMLRAAHTHQTSLQLWNQIIGVNLTGTFLVVREALPALLDNARSAVV NFSSTSASFAHPYMAAYAASKGGIQAFTHSLALEYARRGLRAVCVAPGSIKSGITDATGGYIPEDADWSL FSRLLPVLPTTVESSGTGMAEPTAVAGVIAMLVSDDGAFITGTEIRIDGGTHA
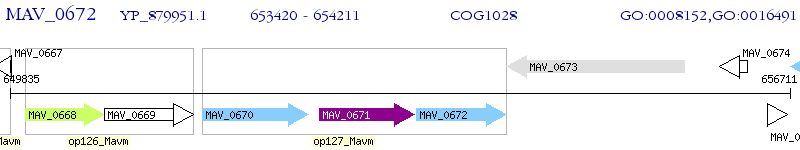
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0672 | - | - | 100% (263) | oxidoreductase |
| M. avium 104 | MAV_3034 | - | 3e-79 | 56.27% (263) | oxidoreductase |
| M. avium 104 | MAV_1572 | - | 1e-32 | 35.14% (259) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1385 | fabG | 3e-30 | 34.50% (258) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. gilvum PYR-GCK | Mflv_4541 | - | 1e-76 | 56.93% (267) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv1350 | fabG | 3e-30 | 34.50% (258) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. leprae Br4923 | MLBr_01740 | - | 2e-21 | 32.81% (192) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4680c | - | 4e-24 | 29.57% (257) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_4967 | - | 1e-116 | 79.85% (263) | short-chain type dehydrogenase/reductase |
| M. smegmatis MC2 155 | MSMEG_2852 | - | 9e-79 | 58.05% (267) | 3-oxoacyl- |
| M. thermoresistible (build 8) | TH_2959 | - | 2e-77 | 58.58% (268) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_4041 | - | 1e-117 | 80.23% (263) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_2848 | - | 2e-76 | 56.13% (269) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1385|M.bovis_AF2122/97 ----------MASLLNARTAVITGGAQGLGLAIGQRFVAEGARVVLGDVN
Rv1350|M.tuberculosis_H37Rv ----------MASLLNARTAVITGGAQGLGLAIGQRFVAEGARVVLGDVN
MAB_4680c|M.abscessus_ATCC_199 -------MNGNSALLQGKNVIVTGGARGLGAAFSRHIVSQGGRVVIADVL
MMAR_4967|M.marinum_M -----------MTRLVGQRILVTGAGSGIGQATVLRLLDEGATVVGADID
MUL_4041|M.ulcerans_Agy99 -----------MTRLVGQRILVTGAGSGIGQATVLRLLDEGATVVGADID
MAV_0672|M.avium_104 -----------MKRLEGKRILVTGAASGIGQATALRLLDEGAAVAASDVA
Mflv_4541|M.gilvum_PYR-GCK ------------MRYSGRRVLITGGGSGIGQACALRILAEGGRVAAADIS
Mvan_2848|M.vanbaalenii_PYR-1 MSATPTGSSPQLTRYAGRRVLITGGGSGIGQACVLRILAEGGRVVAADIS
MSMEG_2852|M.smegmatis_MC2_155 MPDLQAESPIAPSRYAGRRVLITGGGSGIGQACVLRILAEGAQVVAADIS
TH_2959|M.thermoresistible__bu -------VSDLSARYAARRVLITGGGSGIGQACVLRILAEGGRVAAADIS
MLBr_01740|M.leprae_Br4923 -------MVKTAQYFVGKRCFVTGAASGIGRATALRLAAYGAELYLTDRH
.: .:**.. *:* * :: *. : *
Mb1385|M.bovis_AF2122/97 LE---ATEVAAKRLGGDDVALAVRCDVTQADDVDILIRTAVERFGGLDVM
Rv1350|M.tuberculosis_H37Rv LE---ATEVAAKRLGGDDVALAVRCDVTQADDVDILIRTAVERFGGLDVM
MAB_4680c|M.abscessus_ATCC_199 DG---DGRQLAAELG--DAARFTHLDVTDAEQWRELIDTTLAGFGTITGL
MMAR_4967|M.marinum_M AAGLSRTQARTAEAGTGARLTTCQIDVADEKSVVDGVGAAVQALGGLDAL
MUL_4041|M.ulcerans_Agy99 ADGLSRTQARTAEAGTGARLTTCQIDVADEKSVVDGVGAAVQALGGLDAL
MAV_0672|M.avium_104 VDGLERTRDRAAQAGTAERLVTVTMDVGREDAVVDGVRVAVEQLGGLDSL
Mflv_4541|M.gilvum_PYR-GCK GDGLADTVQKAGDLAT--NLMTVVMDVGDEPSVSSGVAEAIAALGGLDTL
Mvan_2848|M.vanbaalenii_PYR-1 ADGLADTAEKAGALGT--GLSTVVMDVGDEQSVSAGLAEALNTLGGLDTL
MSMEG_2852|M.smegmatis_MC2_155 PEGLDDTTGKAGEFSS--RLTTVTLDVGDEDSVRAGVAAAVAELGGLDTL
TH_2959|M.thermoresistible__bu PSGLRDTVNQAGAAAE--RLTTVTMDVADEQSVRHGVGQAITALGGLDAL
MLBr_01740|M.leprae_Br4923 CDGLAQTVSEARALGA-QVPEYRALDISDYDEVAAFGADIHARHPSMDIV
: . *: : :
Mb1385|M.bovis_AF2122/97 VNNAGITRDATMRTMTEEQFDQVIAVHLKGTWNGTRLAAAIMRERKRG-A
Rv1350|M.tuberculosis_H37Rv VNNAGITRDATMRTMTEEQFDQVIAVHLKGTWNGTRLAAAIMRERKRG-A
MAB_4680c|M.abscessus_ATCC_199 VNNAGISSAAMVADEPLEHFRKVIEINLVGVFIGMQAVIPAMRAHGCG-S
MMAR_4967|M.marinum_M VNAAGMLRAAHTHQTSIQQWNQIIAVNLTGTFLVVREALPALLDNQRS-T
MUL_4041|M.ulcerans_Agy99 VNAAGMLRAAHTHQTSIQQWNQIIAVNLTGTFLVVREALPALLDNQRS-T
MAV_0672|M.avium_104 VNAAGMLRAAHTHQTSLQLWNQIIGVNLTGTFLVVREALPALLDNARS-A
Mflv_4541|M.gilvum_PYR-GCK VNAAGILRSAHLADTTLADFEQVLRINLIGTFLVTRESLAALRDGRGP-A
Mvan_2848|M.vanbaalenii_PYR-1 VNAAGILRSAHLAQTTLADFEQVLRINLIGTFLVTREAVTALHQGDGS-A
MSMEG_2852|M.smegmatis_MC2_155 VNAAGILRSSHLAQTSLADFEQVLRINLTGTFLVTREAIGALLEGHRP-A
TH_2959|M.thermoresistible__bu VNAAGILRSAHLVQTTLADFEQVIRINLTGTFLVTRECIPALLDGNGP-A
MLBr_01740|M.leprae_Br4923 MNIAGVSVWGTVDRLTHDQWSNVVAINLMGPIHIIETFVPAILAAGRGGH
:* **: . . : ::: ::* * . :
Mb1385|M.bovis_AF2122/97 IVNMSSVSGKVGMVGQTNYSAAKAGIVGMTKAAAKELAHLGIRVNAIAPG
Rv1350|M.tuberculosis_H37Rv IVNMSSVSGKVGMVGQTNYSAAKAGIVGMTKAAAKELAHLGIRVNAIAPG
MAB_4680c|M.abscessus_ATCC_199 IVNISSAAGLVGLALTSGYGASKWGVRGLSKVAAIELGRERIRVNSVHPG
MMAR_4967|M.marinum_M VVNFSSTSAAFAHPYMAAYAASKGGVQAFTHSLALEYARDGLRAVCVAPG
MUL_4041|M.ulcerans_Agy99 VVNFSSTSAAFAHPYMAAYAASKGGVQAFTHSLALEYARDGLRAVCVAPG
MAV_0672|M.avium_104 VVNFSSTSASFAHPYMAAYAASKGGIQAFTHSLALEYARRGLRAVCVAPG
Mflv_4541|M.gilvum_PYR-GCK VVNFSSTSAHFAHPYMAAYAASKGGVLAMTHAWALEFAKAGIRFNCVQPG
Mvan_2848|M.vanbaalenii_PYR-1 VVNFSSTSAQFAHPYMAAYAASKGGVLAMTHAWALEFAKAGIRFNCVQPG
MSMEG_2852|M.smegmatis_MC2_155 VVNFSSTSAHFAHPYMAAYAASKGGVLAMTHAWALEFAKAGIRFNSVQPG
TH_2959|M.thermoresistible__bu VVNFSSTSAHFAHPYMAAYAASKGGVLAMTHTWAAEFAKAGIRFNAVQPG
MLBr_01740|M.leprae_Br4923 LVNVSSAAGLVALPWHAAYSASKYGLRGLSEVLRFDLARYRIGVSVVVPG
:**.**.:. .. : *.*:* *: .::. : .: : : **
Mb1385|M.bovis_AF2122/97 LIRSAMT---------------EAMPQRIWDQKLAEVP------MGRAGE
Rv1350|M.tuberculosis_H37Rv LIRSAMT---------------EAMPQRIWDQKLAEVP------MGRAGE
MAB_4680c|M.abscessus_ATCC_199 VIYTPMT---------------AKEGFREGEGNMPIAA------MGRVGN
MMAR_4967|M.marinum_M SIKSGITDATGG-------YIPADANWSLFTRLMPILPTTETSSGAGMAD
MUL_4041|M.ulcerans_Agy99 SIKSGITDATGG-------YIPADANWSLFTRLMPILPTTETSSGAGMAD
MAV_0672|M.avium_104 SIKSGITDATGG-------YIPEDADWSLFSRLLPVLPTTVESSGTGMAE
Mflv_4541|M.gilvum_PYR-GCK SISSGMTDGTGASRQSVGPGLPEDADYALFGKVAPMLP---LDDGAIFAA
Mvan_2848|M.vanbaalenii_PYR-1 SISSGMTDGTGASRQSIGPGLPEDADYTLFAKVAPMLP---LDGGAIFAA
MSMEG_2852|M.smegmatis_MC2_155 SISSGMTDGTGASRQSIGPGLPEDADFSLFGKVAPMLP---LDGGAIFAR
TH_2959|M.thermoresistible__bu SISSGMTDGSGVSRQSVGPGLPEDADLALFAKVAPALP---LGDGTIFAD
MLBr_01740|M.leprae_Br4923 AVRTPLVDTIEIAG--------VDRQDPRFSRWVDRFR------GHAVSA
: : :. .
Mb1385|M.bovis_AF2122/97 PSEVASVAVFLASDLSSYMTGTVLDVTGGRFI------------------
Rv1350|M.tuberculosis_H37Rv PSEVASVAVFLASDLSSYMTGTVLDVTGGRFI------------------
MAB_4680c|M.abscessus_ATCC_199 ADEVAGAVAYLLSDAASYVTGAELAVDGGWTAGPTLTTITGQ--------
MMAR_4967|M.marinum_M PSVVAGVIAMLVSEDGAFITGTEIRIDGGTHA------------------
MUL_4041|M.ulcerans_Agy99 PSVVAGVIAMLVSEDGAFITGTEIRIDGGTHA------------------
MAV_0672|M.avium_104 PTAVAGVIAMLVSDDGAFITGTEIRIDGGTHA------------------
Mflv_4541|M.gilvum_PYR-GCK PDAVAGVVAMLGSSDAYFITGTEVRIDGGSHM------------------
Mvan_2848|M.vanbaalenii_PYR-1 PDAVAGVVAMLGSRDAYFITGTEVRIDGGSHM------------------
MSMEG_2852|M.smegmatis_MC2_155 PDAVAGVVAMLGSADAYFITGTEVRIDGGSHM------------------
TH_2959|M.thermoresistible__bu PDAVAGVVAMLGSDDAFFITGTEVRIDGGAHM------------------
MLBr_01740|M.leprae_Br4923 ERAAEKILAGVAKNRYLIYTSHDIRVLYGFKRLAWWPYGVVMKQVNVVFT
. . : . *. : : *
Mb1385|M.bovis_AF2122/97 ----------------------------------
Rv1350|M.tuberculosis_H37Rv ----------------------------------
MAB_4680c|M.abscessus_ATCC_199 ----------------------------------
MMAR_4967|M.marinum_M ----------------------------------
MUL_4041|M.ulcerans_Agy99 ----------------------------------
MAV_0672|M.avium_104 ----------------------------------
Mflv_4541|M.gilvum_PYR-GCK ----------------------------------
Mvan_2848|M.vanbaalenii_PYR-1 ----------------------------------
MSMEG_2852|M.smegmatis_MC2_155 ----------------------------------
TH_2959|M.thermoresistible__bu ----------------------------------
MLBr_01740|M.leprae_Br4923 RALRPSPATMRCVEPIKVGVYSQPRSGEGGKSGN