For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GMSQPTHEMFEAAYRGESPEMGEGARPPWSIGEPQPEIAALIEAGKFHGDVLDAGCGEAAVSLYLAERGF TTVGLDQSPTAIKLAREKAARRGLTSASFEVADISDFTGYDGRFGTIVDSTLFHSMPVELREGYQRSIVQ AAAPGASYFVLVFDRNAMPAEGPVNAVTEDELREVVGKYWVIDEIRPARIHANVPENFLAAFEAFAGADI RDEDNGRKSVGAWLLSAHLG*
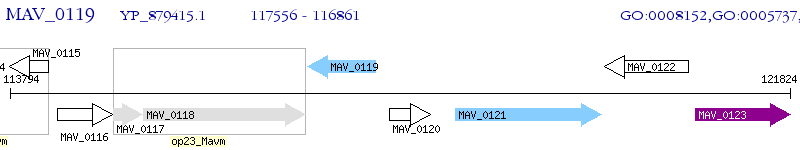
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0119 | - | - | 100% (231) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. avium 104 | MAV_0404 | - | 1e-64 | 60.63% (221) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. avium 104 | MAV_0313 | - | 7e-47 | 45.42% (240) | thiopurine S-methyltransferase (tpmt) superfamily protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0575c | - | 4e-89 | 73.76% (221) | benzoquinone methyltransferase |
| M. gilvum PYR-GCK | Mflv_1324 | - | 2e-56 | 54.71% (223) | methyltransferase type 11 |
| M. tuberculosis H37Rv | Rv0560c | - | 4e-89 | 73.76% (221) | benzoquinone methyltransferase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0393 | - | 1e-83 | 71.82% (220) | methyltransferase |
| M. marinum M | MMAR_0909 | - | 4e-88 | 69.53% (233) | methyltransferase |
| M. smegmatis MC2 155 | MSMEG_2539 | - | 9e-98 | 77.13% (223) | thiopurine S-methyltransferase (tpmt) superfamily protein |
| M. thermoresistible (build 8) | TH_1722 | - | 7e-64 | 56.22% (233) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0662 | - | 6e-75 | 68.69% (198) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_5471 | - | 5e-59 | 55.86% (222) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_0119|M.avium_104 --------------MGMSQPTHEMFEAAYRGESPEMGEGARPPWSIGEPQ
MSMEG_2539|M.smegmatis_MC2_155 ----------------MTTP-EDLFESAYRGEAPNFG-GVRPPWSIGEPQ
Mb0575c|M.bovis_AF2122/97 MSTVLTYIRAVDIYEHMTESLDLEFESAYRGESVAFGEGVRPPWSIGEPQ
Rv0560c|M.tuberculosis_H37Rv MSTVLTYIRAVDIYEHMTESLDLEFESAYRGESVAFGEGVRPPWSIGEPQ
MAB_0393|M.abscessus_ATCC_1997 -----------MTDKTATEEFDLAFDAAYRGEAPDGR-GARPPWSIGEPQ
MMAR_0909|M.marinum_M ----------------MTESLDLQFETAYRGESAEFGAGIRPPWSLGEPQ
MUL_0662|M.ulcerans_Agy99 ----------------MTESLDLQFATAYRGESAEFGAGIRPPWSLGEPQ
Mflv_1324|M.gilvum_PYR-GCK ------------------MAEVMDWDSAYRGEGE-FE--GEPPWNIGEPQ
Mvan_5471|M.vanbaalenii_PYR-1 ---------------MAHMTDVMDWDSAYRGEGD-FE--GEPPWNIGEPQ
TH_1722|M.thermoresistible__bu --------MATEDPGTGDPTDTIDWDSVYRQEAPGFD--GPPPWNIGEPQ
: :.** *. ***.:****
MAV_0119|M.avium_104 PEIAALIEAGKFHGDVLDAGCGEAAVSLYLAERGFTTVGLDQSPTAIKLA
MSMEG_2539|M.smegmatis_MC2_155 PEIRKLIDAGKFHGEVLDAGCGEAATALYLAAQGFTTVGLDQSPTAIELA
Mb0575c|M.bovis_AF2122/97 PELAALIVQGKFRGDVLDVGCGEAAISLALAERGHTTVGLDLSPAAVELA
Rv0560c|M.tuberculosis_H37Rv PELAALIVQGKFRGDVLDVGCGEAAISLALAERGHTTVGLDLSPAAVELA
MAB_0393|M.abscessus_ATCC_1997 PEIAMLIAEGKVRGEVLDAGCGEAATSLYLAEVGHTTVGLDSAPKAIELA
MMAR_0909|M.marinum_M PELATLIEQGKVHGEVLDAGCGEAALALALAGRGHPVVGLDMSPTAVELA
MUL_0662|M.ulcerans_Agy99 PELATLIEQGKVHGEVLDAGCGEAALAMALAGRGHPVVGLDISPTAVELA
Mflv_1324|M.gilvum_PYR-GCK PELAALWRDGRFRSEVLDAGCGHAELSLALAADGYTVTGVDISPTAMAAA
Mvan_5471|M.vanbaalenii_PYR-1 PELAALWRDGKFRSEVLDAGCGHAELSLALAADGYTVTGVDLSPTAVAAA
TH_1722|M.thermoresistible__bu PELAALHRAGAFRSDVLDAGCGHAEISMALAADGYTVVGIDLSPTAIAAA
**: * * .:.:***.***.* :: ** *....*:* :* *: *
MAV_0119|M.avium_104 REKAARRGLTS-ASFEVADISDFTGY----DGRFGTIVDSTLFHSMPVEL
MSMEG_2539|M.smegmatis_MC2_155 RAEAARRGLTN-ATFEVADISSFTGY----DGRFGTIVDSTLFHSMPVEL
Mb0575c|M.bovis_AF2122/97 RHEAAKRGLAN-ASFEVADASSFTGY----DGRFDTIVDSTLFHSMPVES
Rv0560c|M.tuberculosis_H37Rv RHEAAKRGLAN-ASFEVADASSFTGY----DGRFDTIVDSTLFHSMPVES
MAB_0393|M.abscessus_ATCC_1997 KGYAAERGLTN-ASFAVADISSFTGY----DGRFDTIIDSTLFHSMPVGL
MMAR_0909|M.marinum_M GREAARRGLTN-ASFAVADITDFASYPPESAGRFNTIMDSTLFHSMPVEL
MUL_0662|M.ulcerans_Agy99 GREAARPGLTN-ASFAVADITDFASYPPESAGRFNTIMDSTLFHSMPVEL
Mflv_1324|M.gilvum_PYR-GCK SAAAAERGLSEKASFVAADITALSGF----DGRFSTVVDSTLFHSLPVES
Mvan_5471|M.vanbaalenii_PYR-1 TAASAERGLSERTSFVAADITTLTGF----DGRFATVVDSTLFHSLPVES
TH_1722|M.thermoresistible__bu NRTAQERGLAT-ASFVQGDITDFGGY----DGRFNTIIDSTLFHSIPVQA
: . **: ::* .* : : .: *** *::*******:**
MAV_0119|M.avium_104 REGYQRSIVQAAAPGASYFVLVFDRNAMPAEGPVNA----VTEDELREVV
MSMEG_2539|M.smegmatis_MC2_155 RDGYQQSIVRAAAPGATYVVLVFDKTTMGDTGPANP----VTDDELRDVV
Mb0575c|M.bovis_AF2122/97 REGYLQSIVRAAAPGASYFVLVFDRAAIP-EGPINA----VTEDELRAAV
Rv0560c|M.tuberculosis_H37Rv REGYLQSIVRAAAPGASYFVLVFDRAAIP-EGPINA----VTEDELRAAV
MAB_0393|M.abscessus_ATCC_1997 RAGYQRSIVRAAAPGARYYVLVFDRAAFPAESPVNA----VTENELRDAV
MMAR_0909|M.marinum_M REGYQRSIVRAAAPGASYFVLVFDKAALSGDGPING----VTEEELRDAV
MUL_0662|M.ulcerans_Agy99 REGCQRSIVRAAAPGASYFVLVFDKAALSGDGPING----VTEEELRDAV
Mflv_1324|M.gilvum_PYR-GCK RAGYLESVHRAARPGAGFFVLVFAKGAFPADTEQGP--NEVSEIELREAV
Mvan_5471|M.vanbaalenii_PYR-1 RDGYLQSVHRAAAPGAGYFVLVFAKGAFPAEMEGGP--NEVTELELREAV
TH_1722|M.thermoresistible__bu RDGYLRSIHRAAAPGARYFALVFAKGAFPEEMEQGPGPNPVDEDELREAV
* * .*: :** *** : .*** : :: . * : *** .*
MAV_0119|M.avium_104 GKYWVIDEIRPARIHANVPENFLAAFEAFAGADIRDEDNGRKSVG-----
MSMEG_2539|M.smegmatis_MC2_155 GRYWVIDDISPARIHANVPAEFT-DFAAFAGSDIRDEGNGRKSVA-----
Mb0575c|M.bovis_AF2122/97 SKYWIIDEIKPARLYARFPAGFA-GMPALL--DIREEPNGLQSIG-----
Rv0560c|M.tuberculosis_H37Rv SKYWIIDEIKPARLYARFPAGFA-GMPALL--DIREEPNGLQSIG-----
MAB_0393|M.abscessus_ATCC_1997 SKYWAIDEIRPAKIYANFNGFEPGVARGFARHEIEPD--GRASVA-----
MMAR_0909|M.marinum_M SKYWVIDEIRPARIHANLPPEVSGMPAFDARDEP-EGRKSVG--------
MUL_0662|M.ulcerans_Agy99 SKHWVIDEIRPARIHANLPPEVPGCPPSTLATNPRAASPSVRGCSQPIWA
Mflv_1324|M.gilvum_PYR-GCK SPYWTIDDIRPALIHTNVPKLPGMPPPPLDMLDDRGHLRMQ---------
Mvan_5471|M.vanbaalenii_PYR-1 SRYWIIDDIRPALIDTNVPKLPGMPPPPLDMLDDRGRLRMQ---------
TH_1722|M.thermoresistible__bu SKYWEIDEIRPAFIHANMP--PDVPGVESFDTDEKGRTKLP---------
. :* **:* ** : :.. :
MAV_0119|M.avium_104 -----AWLLSAHLG-
MSMEG_2539|M.smegmatis_MC2_155 -----GWLLQAHLG-
Mb0575c|M.bovis_AF2122/97 -----GWLLSAHLG-
Rv0560c|M.tuberculosis_H37Rv -----GWLLSAHLG-
MAB_0393|M.abscessus_ATCC_1997 -----AWLLSAHLAG
MMAR_0909|M.marinum_M -----AWLLSAHLG-
MUL_0662|M.ulcerans_Agy99 DGTPHAVYSPANEG-
Mflv_1324|M.gilvum_PYR-GCK -----AFLLSAHKEP
Mvan_5471|M.vanbaalenii_PYR-1 -----AFLLQAHKES
TH_1722|M.thermoresistible__bu -----AFLLSAHKA-
. *: