For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SAIRVLVADDDTLLREGVASVLEKAGFEVVGRAGDATALLELVRTESPNLVVVDIRMPPTHTTEGLEAAQ KIRGEHPGIGVLVLSAHVEVEYAAELITSGDGVGYLLKSRIGDVGEFADACTRVAGGGSIVDPELVRELL SARRKDDPLSALSQRELEVLELMAQGLSNSGIAGRLWITESTVEKHVHSVMRRLDLGHADEYHRRVLAVL AFLTHR*
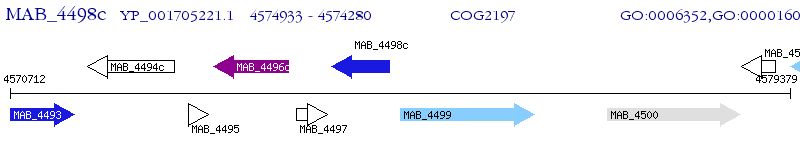
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4498c | - | - | 100% (217) | two-component LuxR family response regulator |
| M. abscessus ATCC 19977 | MAB_4519c | - | 2e-23 | 35.71% (210) | two-component LuxR family response regulator |
| M. abscessus ATCC 19977 | MAB_3891c | - | 7e-22 | 31.31% (214) | LuxR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0867c | narL | 5e-20 | 35.38% (195) | nitrate/nitrite response transcriptional regulatory protein |
| M. gilvum PYR-GCK | Mflv_2280 | - | 1e-48 | 47.17% (212) | two component LuxR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0844c | narL | 1e-20 | 35.90% (195) | nitrate/nitrite response transcriptional regulatory protein |
| M. leprae Br4923 | MLBr_01286 | - | 1e-10 | 31.43% (105) | putative two-component response regulatory protein |
| M. marinum M | MMAR_4782 | narL | 1e-22 | 36.92% (195) | nitrate/nitrite response regulator protein NarL |
| M. avium 104 | MAV_5280 | - | 2e-20 | 33.33% (201) | two-component system response regulator |
| M. smegmatis MC2 155 | MSMEG_0983 | - | 1e-74 | 62.67% (217) | two-component system response regulator |
| M. thermoresistible (build 8) | TH_1532 | - | 4e-48 | 47.64% (212) | two-component system response regulator |
| M. ulcerans Agy99 | MUL_0351 | narL | 2e-22 | 36.92% (195) | nitrate/nitrite response regulator protein NarL |
| M. vanbaalenii PYR-1 | Mvan_2073 | - | 6e-74 | 61.29% (217) | two component LuxR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0983|M.smegmatis_MC2_155 ----------MSGLQVVVADDDVLLREGLSSLLDRSG-FDVVGQAADATE
Mvan_2073|M.vanbaalenii_PYR-1 ----------MSALKVVVADDDVLLREGLASLLERSG-FDVVGQAGDGEK
MAB_4498c|M.abscessus_ATCC_199 ----------MSAIRVLVADDDTLLREGVASVLEKAG-FEVVGRAGDATA
Mflv_2280|M.gilvum_PYR-GCK -------------MRIVIAEDSALLRAGIERILTDAG-HQVVAGVPDATD
TH_1532|M.thermoresistible__bu -------------MRIVIAEDSALLRAGIERILTDAG-HEVVAGVPDATD
MMAR_4782|M.marinum_M -----MDVTKTEKVRVVVGDDHPLFREGVVRALSLSGSVNVVGEADDGAA
MUL_0351|M.ulcerans_Agy99 -----MDVTKTEKVRVVVGDDHPLFREGVVRALSLSGSVNVVGEADDGAA
Mb0867c|M.bovis_AF2122/97 -----MSNPQPEKVRVVVGDDHPLFREGVVRALSLSGSVNVVGEADDGAA
Rv0844c|M.tuberculosis_H37Rv -----MSNPQPEKVRVVVGDDHPLFREGVVRALSLSGSVNVVGEADDGAA
MAV_5280|M.avium_104 -----MTDPAP-EITVLLVDDQDLVRSGLRRILRRKDGFVIVAECADGDE
MLBr_01286|M.leprae_Br4923 MTGPTTDADAATPHRVLIAEDEALIRLDLAEMLRDEG-YDIVGEAADGQE
::: :* *.* .: * . :*. *.
MSMEG_0983|M.smegmatis_MC2_155 LLTLVREKTPQLAVVDIRMPPTNTTEGLDAAKVIRSEFPDTGILVLSAHV
Mvan_2073|M.vanbaalenii_PYR-1 LLELVRTRQPALVLTDIRMPPTHTSEGLDAAKIIRQESPEVGIVVLSAHV
MAB_4498c|M.abscessus_ATCC_199 LLELVRTESPNLVVVDIRMPPTHTTEGLEAAQKIRGEHPGIGVLVLSAHV
Mflv_2280|M.gilvum_PYR-GCK LLRLVNDERPDLAILDVRMPPTFTDEGIRAAALLRSQNPESPVLVLSHYV
TH_1532|M.thermoresistible__bu LLRLVNELRPDLAIVDVRMPPTFTDEGIRAAALLRSQNPESPVLVLSHYV
MMAR_4782|M.marinum_M ALELIKTHLPDVALLDYRMP---RMDGAQVAAAVRSNELPTRVLLISAHD
MUL_0351|M.ulcerans_Agy99 ALELIETHLPDVALLDYRMP---RMDGAQVAAAVRSNGLPTRVLLISAHD
Mb0867c|M.bovis_AF2122/97 ALELIKAHLPDVALLDYRMP---GMDGAQVPAAVRSYELPTRVLLISAHD
Rv0844c|M.tuberculosis_H37Rv ALELIKAHLPDVALLDYRMP---GMDGAQVAAAVRSYELPTRVLLISAHD
MAV_5280|M.avium_104 VPAAVAAHRPDVVVMDLRMR---RVDGIEATRRLGGR---PPVLALTTFN
MLBr_01286|M.leprae_Br4923 AVELAERHKPDLVIMDVKMP---RRDGIDAASEIASKR-IAPIVVLTAFS
* :.: * :* :* .. : :: :: .
MSMEG_0983|M.smegmatis_MC2_155 EVEHATELLASGH-GIGYLLKSRVTDVSDFVETLERIAKGASVVDPALVS
Mvan_2073|M.vanbaalenii_PYR-1 DVDHAMELLAGGH-AIGYLLKTRVTDVADFVDTLQRIANGASVVDPALVA
MAB_4498c|M.abscessus_ATCC_199 EVEYAAELITSGD-GVGYLLKSRIGDVGEFADACTRVAGGGSIVDPELVR
Mflv_2280|M.gilvum_PYR-GCK EERYAADLIASDTRGFGYLLKDRVADVPAFLDAVEVVGAGGTVLDPEVVS
TH_1532|M.thermoresistible__bu EERYAADLIASDTKGFGYLLKDRVADVPAFLDAVEVVGGGGTVLDPEVVS
MMAR_4782|M.marinum_M ESAIVYQALQQGA--AGFLLKD--STRTEIVKAVLDCAKGRDVVAPSLVG
MUL_0351|M.ulcerans_Agy99 ESAIVYQALQQGA--AGFLLKD--STRTEIVKAVLDCAKGRDVVAPSLVG
Mb0867c|M.bovis_AF2122/97 EPAIVYQALQQGA--AGFLLKD--STRTEIVKAVLDCAKGRDVVAPSLVG
Rv0844c|M.tuberculosis_H37Rv EPAIVYQALQQGA--AGFLLKD--STRTEIVKAVLDCAKGRDVVAPSLVG
MAV_5280|M.avium_104 EDELLSGALRAGA--AGFVLKD--SSAEELIRAVRAVARGEGYLDPAVTS
MLBr_01286|M.leprae_Br4923 QRDLVERARDAGA--MAYLVKP--FTISDLIPAIALAMSRFSELT-ALER
: . .:::* : : : :
MSMEG_0983|M.smegmatis_MC2_155 ELVSARRRDDPL------AALSAREQEVLNLMAEGRSNAGIARRLWVTEG
Mvan_2073|M.vanbaalenii_PYR-1 ELVSARKRDDPL------AALSAREHEVLTLMAEGLSNGGIGRRLWVTEG
MAB_4498c|M.abscessus_ATCC_199 ELLSARRKDDPL------SALSQRELEVLELMAQGLSNSGIAGRLWITES
Mflv_2280|M.gilvum_PYR-GCK QILARSHRRNVL------DALTPRENEVLQLMAEGKTNSAIASGLHMSVG
TH_1532|M.thermoresistible__bu QILVRSHRRSAL------DELTAREHDVLRLMAEGKTNSAIAGALHISIG
MMAR_4782|M.marinum_M GLAGEIRQRAAP----MAPVLSAREREVLHRIAKGQSIPAIAGELYVAPS
MUL_0351|M.ulcerans_Agy99 GLAGEIRQRAAP----MAPVLSAREREVLHRIAKGQSIPAIAGELYVAPS
Mb0867c|M.bovis_AF2122/97 GLAGEIRQRAAP----VAPVLSAREREVLNRIACGQSIPAIAAELYVAPS
Rv0844c|M.tuberculosis_H37Rv GLAGEIRQRAAP----VAPVLSAREREVLNRIACGQSIPAIAAELYVAPS
MAV_5280|M.avium_104 RVLTTYRKAAPGPHGAAIAELTTRERDVLTLIGKGLSNSEIADELCISGV
MLBr_01286|M.leprae_Br4923 EVATLADRLETR-------KLVERAKGLLQ-VKQGMTEPEAFKWIQRAAM
: : * * :* : * : : :
MSMEG_0983|M.smegmatis_MC2_155 TVEKHVRSILTKLNLTETADDHRRVRAVITYLESR
Mvan_2073|M.vanbaalenii_PYR-1 TVEKHVRSILTKLNLPETGDDHRRVRAVIMYLETR
MAB_4498c|M.abscessus_ATCC_199 TVEKHVHSVMRRLDLGHADEYHRRVLAVLAFLTHR
Mflv_2280|M.gilvum_PYR-GCK SAEKHIASIFTKLGLAPDDTENRRVLAVLRYLES-
TH_1532|M.thermoresistible__bu SAEKHIASIFTKLGLEPDESENRRVLAVLRYLES-
MMAR_4782|M.marinum_M TVKTHVQRLYEKLGVSDRAAAVAEAMRQGLLD---
MUL_0351|M.ulcerans_Agy99 TVKTHVQRLYEKLGVSDRAAAVAEAMRQGLLD---
Mb0867c|M.bovis_AF2122/97 TVKTHVQRLYEKLGVSDRAAAVAEAMRQRLLD---
Rv0844c|M.tuberculosis_H37Rv TVKTHVQRLYEKLGVSDRAAAVAEAMRQRLLD---
MAV_5280|M.avium_104 TVKSHIGRIFGKLDLRDRAAAIVYAYDNGIVAPR-
MLBr_01286|M.leprae_Br4923 DRRTTMKRVAEVVLETLDTPKDT------------
.. : : :