For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TNDIRPVCVVGGGRMGAGISQVFALAGFTVFVYEVNEQVRTSVVDRVRESLACAELDVAVADRIIITDDL SAAVADACFVTEAVGEKLELKREVLAGIAAAAPHDAILASNTSVIPISEIAAGSPAIDRIVGTHWWNPAP LIPLVEVVQATGTSVETVSRTMKLLAAIGKKPMHVRKEVPGFVGNRLQHALWREAIALVQEGVCDATTVD DVVKNSFGLRLAVLGPLENADLVGLDLTLDIHRVVVPALDCQGGPNPVLVDKVARGMLGCKTGEGFRRWS PADIAEMNNRLEGHLVRLLHPRNTGPVG*
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
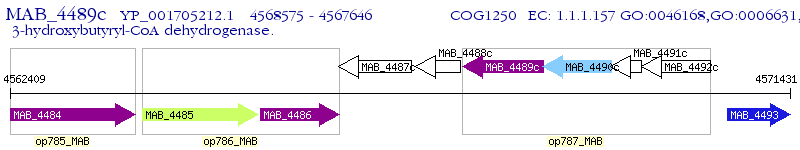
| M. abscessus ATCC 19977 | MAB_4489c | - | - | 100% (309) | 3-hydroxyacyl-CoA dehydrogenase family protein |
| M. abscessus ATCC 19977 | MAB_0904 | - | 3e-44 | 40.21% (281) | putative 3-hydroxyacyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4094c | - | 4e-32 | 36.11% (288) | 3-hydroxybutyryl-CoA dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1742 | fadB3a | 7e-34 | 42.62% (183) | 3-hydroxybutyrul-CoA dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0106 | - | 6e-32 | 34.86% (284) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. tuberculosis H37Rv | Rv1715 | fadB3 | 6e-68 | 46.80% (297) | 3-hydroxybutyryl-CoA dehydrogenase FADB3 |
| M. leprae Br4923 | MLBr_02461 | fadB2 | 6e-32 | 34.98% (283) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. marinum M | MMAR_0793 | fadB2 | 1e-32 | 36.62% (284) | 3-hydroxybutyryl-CoA dehydrogenase FadB2 |
| M. avium 104 | MAV_4681 | - | 5e-31 | 35.69% (283) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6791 | - | 8e-42 | 34.07% (317) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_3443 | - | 3e-27 | 34.51% (255) | PUTATIVE 3-hydroxybutyryl-CoA dehydrogenase FadB2 |
| M. ulcerans Agy99 | MUL_4537 | fadB2 | 2e-32 | 36.27% (284) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0802 | - | 3e-32 | 35.56% (284) | 3-hydroxybutyryl-CoA dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0793|M.marinum_M -----MG----SDAAIQRVGVVGAGQMGSGIAEVSVRAGVEVTVFETTEA
MUL_4537|M.ulcerans_Agy99 -----MG----SDAAIQRVGVVGAGQMGSGIAEVSVRAGVEVTAFETTEA
MAV_4681|M.avium_104 MHDKTIGGETVSDAAIQRVGVVGAGQMGSGIAEVSVRADVDVTVFETTEA
MLBr_02461|M.leprae_Br4923 ----------MSDAAIQRVGVVGAGQMGSGIAEVAVRAGVGVTVFETTDT
TH_3443|M.thermoresistible__bu ------------------------------LRWWHARERCPX-VYDISEE
Mflv_0106|M.gilvum_PYR-GCK -------------MSIERVGVVGAGQMGSGIAEVAAKAGANVVVYEPSDA
Mvan_0802|M.vanbaalenii_PYR-1 -------------MSIERVGVVGAGQMGSGIAEVAAKAGANVVVYEPSEA
Mb1742|M.bovis_AF2122/97 ---------MLTSHGFSRAAVVGAGLMGRRIAGVLASAGLDVAITDTNAE
Rv1715|M.tuberculosis_H37Rv ---------MLTSHGFSRAAVVGAGLMGRRIAGVLASAGLDVAITDTNAE
MAB_4489c|M.abscessus_ATCC_199 -----------MTNDIRPVCVVGGGRMGAGISQVFALAGFTVFVYEVNEQ
MSMEG_6791|M.smegmatis_MC2_155 ------------MRKINTVTVVGAGYMGGGIAQVLALNGFKVQIADVNAE
: . : .
MMAR_0793|M.marinum_M LITAGRNRIVKSLERGVSSGKVTERERDRALSR--LTFTTDLNDLADRQL
MUL_4537|M.ulcerans_Agy99 LITAGRNRIVKSLERGVSSGKVTERERDRALSR--LTFTTDLNDLADRQL
MAV_4681|M.avium_104 LITAGRNRIVKSLERGVSAGKVTERERDRALSK--LTFTTDLKDLADRQL
MLBr_02461|M.leprae_Br4923 LVTAGRNRIVKSLERGVSAGKVTERERDSALGH--LVFTTDLNDLADRQL
TH_3443|M.thermoresistible__bu LTTAGRERIAKSLTRGVQAGKLSEDERSAALDR--LRFTTTLADLSDRQL
Mflv_0106|M.gilvum_PYR-GCK LVNAGRERITASLERAKAKGKLAADDFEITLAR--LTFTTDLADLADRQL
Mvan_0802|M.vanbaalenii_PYR-1 LVAAGRDRITASLERAKAKGKLAADDFDATVAR--LTFTTNLGDLADRQL
Mb1742|M.bovis_AF2122/97 ILHAAAVEAARVA---------GAGRGSVAAAA------DLAAAIPDADL
Rv1715|M.tuberculosis_H37Rv ILHAAAVEAARVA---------GAGRGSVAAAA------DLAAAIPDADL
MAB_4489c|M.abscessus_ATCC_199 VRTSVVDRVRESL---------ACAELDVAVADRIIITDDLSAAVADACF
MSMEG_6791|M.smegmatis_MC2_155 ATQEALKRLDREAREFEQQGLFGPGSADTIMSN-LTASDSLDDAVSDVDY
. . :.*
MMAR_0793|M.marinum_M VIEAIIEDEAVKTKVFADLDRVVTDPDAVLASNTSSIPIMKVAAATKNPR
MUL_4537|M.ulcerans_Agy99 VIEAIIEDEAVKTKVFADLDRVVTDPDAVLASNTSSIPIMKVAAATKNPR
MAV_4681|M.avium_104 VIEAIIEDDAVKAQVFAELDRVVTDPDAVLASNTSSIPIMKLAAATKNPQ
MLBr_02461|M.leprae_Br4923 VIEAIVEDEAVKTKVFAELDRVITDPDAVLASNTSSIPIMKIAAATKNPQ
TH_3443|M.thermoresistible__bu VIEAIVEDENVKATVFAELDELLTDPDAVLASNTSSIPIMKLAAATKNPG
Mflv_0106|M.gilvum_PYR-GCK VIEAIIEDEAVKAKVFAQLDEVVTDPDAVLASNTSSIPIMKIAAATKNPS
Mvan_0802|M.vanbaalenii_PYR-1 VIEAIVENEAVKAKVFAELDAVVTDPDAVLASNTSSIPIMKIAAATKNPS
Mb1742|M.bovis_AF2122/97 VIEAVVENLAVKQELFERLATLAP--DAVLATNTSVLPIGAVTERVEDGS
Rv1715|M.tuberculosis_H37Rv VIEAVVENLAVKQELFERLATLAP--DAVLATNTSVLPIGAVTERVEDGS
MAB_4489c|M.abscessus_ATCC_199 VTEAVGEKLELKREVLAGIAAAAPH-DAILASNTSVIPISEIAAGSPAID
MSMEG_6791|M.smegmatis_MC2_155 VMEAVFEDVDVKKEVLARVCASARP-DTIIGTNTSTIPVKVLVDAVTHPE
* **: *. :* :: : *:::.:*** :*: :.
MMAR_0793|M.marinum_M RVLGLHFFNPVPVLPLVELVSTLVTDEAAVARTEEFASAVLGKQVVRCSD
MUL_4537|M.ulcerans_Agy99 RVLGLHFFNPVPVLPLVELVNTLVTDEAAVARTEEFASAVLGKQVVRCSD
MAV_4681|M.avium_104 RVLGLHFFNPVPVLPLVELVSTLVTDEAAAARTEEFASAVLGKQVVRCSD
MLBr_02461|M.leprae_Br4923 RVLGLHFFNPVPVLPLVELVSTLVTDEAAATRAEEFASGVLGKQVVRCSD
TH_3443|M.thermoresistible__bu RVLGLHFFNPVPVLPLVELINTLVTTDEAIARTEKFAAEVLGKQVVRCGD
Mflv_0106|M.gilvum_PYR-GCK RVLGLHFFNPVPVLPLVEVINTLVTSEEAVARVEQFASEVLGKKVVRCGD
Mvan_0802|M.vanbaalenii_PYR-1 RVLGLHFFNPVPVLPLVELVNTLVTSDDAIARVERFAGEVLGKKVVRCGD
Mb1742|M.bovis_AF2122/97 RVIGTHFWNPPDLIPVVEVVPSARTAPDTADRVVALLTQVGKLPVRVGRD
Rv1715|M.tuberculosis_H37Rv RVIGTHFWNPPDLIPVVEVVPSARTAPDTADRVVALLTQVGKLPVRVGRD
MAB_4489c|M.abscessus_ATCC_199 RIVGTHWWNPAPLIPLVEVVQATGTSVETVSRTMKLLAAIGKKPMHVRKE
MSMEG_6791|M.smegmatis_MC2_155 RFLTVHFSNPAPFIPGVELVAGEATTQEVIDSVKDLLARAGREGAQVA-D
*.: *: ** .:* **:: * . . : :
MMAR_0793|M.marinum_M RSGFVVNALLVPYLLSAIRMVEAGFATVEDVDKAVVAGLSHPMG---PLR
MUL_4537|M.ulcerans_Agy99 RSGFVVNALLVPYLLSAIRMVEAGFATVEDVDKAVVAGLSHPMG---PLR
MAV_4681|M.avium_104 RSGFVVNALLVPYLLSAIRMVEAGFATVEDVDKAVVAGLSHPMG---PLR
MLBr_02461|M.leprae_Br4923 RSGFVVNALLVPYLLSAIRIVEAGFATVEDVDKAVLAGLSHPMG---PLR
TH_3443|M.thermoresistible__bu RSGFVVNALLVPYLLSAIRMVESGYASVEDVDKAVVAGLSHPMG---PLR
Mflv_0106|M.gilvum_PYR-GCK RSGFIVNALLVPYLLSAIRMAEAGVATVEDIDTAVVAGLSHPMG---PLR
Mvan_0802|M.vanbaalenii_PYR-1 RSGFVVNALLVPYLLSAIRMAEAGVATVEDIDTAVVAGLSHPMG---PLR
Mb1742|M.bovis_AF2122/97 VPGFIGNRLQHAL-------------------------------------
Rv1715|M.tuberculosis_H37Rv VPGFIGNRLQHALWREAIALVAEGVCDPKTVDLVVRNTIGLRLATLGPLE
MAB_4489c|M.abscessus_ATCC_199 VPGFVGNRLQHALWREAIALVQEGVCDATTVDDVVKNSFGLRLAVLGPLE
MSMEG_6791|M.smegmatis_MC2_155 TPGMALNRLQYALLKEATLIVEEGVATKEDVDTIVRTTFGFRLGFFGPFA
.*: * * .
MMAR_0793|M.marinum_M LSDLVGLDTLKLIADKMFEEFKEPHYGPPPLLLRMVEAGQLGKKSGQGFY
MUL_4537|M.ulcerans_Agy99 LSDLVGLDTLKLIADKMFEEFKEPHYGPPPLLLRMVEAGQLGKKSGQGFY
MAV_4681|M.avium_104 LSDLVGLDTLKLIADKMFEEFKEPQYGAPPLLLRMVEAGRLGKKTGQGFY
MLBr_02461|M.leprae_Br4923 LSDLVGLDTLKLIADKMFEEFKEPHYGPPPLLLRMVEAGLLGKKSGQGFY
TH_3443|M.thermoresistible__bu LSDLVGLDTLKLIADKMFDEFKEPHYGPPPLLVRMVEAGHLGKKTGRGFY
Mflv_0106|M.gilvum_PYR-GCK LSDLIGLDTMKLIADSMYDEYKDPNFAPPPLLQRMVEAGQLGKKSGKGFY
Mvan_0802|M.vanbaalenii_PYR-1 LSDLIGLDTMKLIADSMYDEYKDPHYAPPPLLQRMVEAGQLGKKSGKGFY
Mb1742|M.bovis_AF2122/97 --------------------------------------------------
Rv1715|M.tuberculosis_H37Rv NADYIGLDLTLAIHDAVIPSLN-HDPHPSPLLRELVAAGQLGARTGHGFL
MAB_4489c|M.abscessus_ATCC_199 NADLVGLDLTLDIHRVVVPALD-CQGGPNPVLVDKVARGMLGCKTGEGFR
MSMEG_6791|M.smegmatis_MC2_155 IADQAGLDVYVKGYHTLENAFG-ERMATPKLLTDNVDAGRYGTKNGKGWT
MMAR_0793|M.marinum_M TY----------------------------------
MUL_4537|M.ulcerans_Agy99 TY----------------------------------
MAV_4681|M.avium_104 TY----------------------------------
MLBr_02461|M.leprae_Br4923 TY----------------------------------
TH_3443|M.thermoresistible__bu TY----------------------------------
Mflv_0106|M.gilvum_PYR-GCK SY----------------------------------
Mvan_0802|M.vanbaalenii_PYR-1 TY----------------------------------
Mb1742|M.bovis_AF2122/97 ------------------------------------
Rv1715|M.tuberculosis_H37Rv ----DWPAGAREATTARLAQHIAAQLQANEKGRGT-
MAB_4489c|M.abscessus_ATCC_199 ----RWSPADIAEMNNRLEGHLVRLLHPRNTGPVG-
MSMEG_6791|M.smegmatis_MC2_155 GDFDDETKAAVIAYRNKAYSRMGELLRELGPAPKGA