For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLTSHGFSRAAVVGAGLMGRRIAGVLASAGLDVAITDTNAEILHAAAVEAARVAGAGRGSVAAAADLAAA IPDADLVIEAVVENLAVKQELFERLATLAPDAVLATNTSVLPIGAVTERVEDGSRVIGTHFWNPPDLIPV VEVVPSARTAPDTADRVVALLTQVGKLPVRVGRDVPGFIGNRLQHALWREAIALVAEGVCDPKTVDLVVR NTIGLRLATLGPLENADYIGLDLTLAIHDAVIPSLNHDPHPSPLLRELVAAGQLGARTGHGFLDWPAGAR EATTARLAQHIAAQLQANEKGRGT
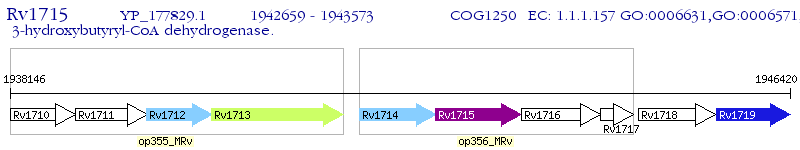
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1715 | fadB3 | - | 100% (304) | PROBABLE 3-HYDROXYBUTYRYL-CoA DEHYDROGENASE FADB3 (BETA-HYDROXYBUTYRYL-CoA DEHYDROGENASE) (BHBD) |
| M. tuberculosis H37Rv | Rv0468 | fadB2 | 2e-32 | 37.85% (288) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. tuberculosis H37Rv | Rv0860 | fadB | 5e-27 | 31.06% (293) | fatty oxidation protein FadB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1742 | fadB3a | 1e-99 | 100.00% (187) | 3-hydroxybutyrul-CoA dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0106 | - | 1e-34 | 37.06% (286) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. leprae Br4923 | MLBr_02461 | fadB2 | 2e-30 | 35.69% (283) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_4489c | - | 6e-68 | 46.80% (297) | 3-hydroxyacyl-CoA dehydrogenase family protein |
| M. marinum M | MMAR_1322 | fadB3 | 1e-32 | 35.51% (276) | 3-hydroxybutyryl-CoA dehydrogenase FadB3 |
| M. avium 104 | MAV_4681 | - | 5e-29 | 34.52% (281) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6791 | - | 7e-38 | 37.32% (276) | 3-hydroxybutyryl-CoA dehydrogenase |
| M. thermoresistible (build 8) | TH_2033 | fadB | 2e-26 | 29.45% (292) | PROBABLE FATTY OXIDATION PROTEIN FADB |
| M. ulcerans Agy99 | MUL_2558 | fadB3 | 8e-33 | 35.51% (276) | 3-hydroxybutyryl-CoA dehydrogenase FadB3 |
| M. vanbaalenii PYR-1 | Mvan_0802 | - | 3e-35 | 39.01% (282) | 3-hydroxybutyryl-CoA dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1715|M.tuberculosis_H37Rv --------------------------------------------------
Mb1742|M.bovis_AF2122/97 --------------------------------------------------
MAB_4489c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1322|M.marinum_M --------------------------------------------------
MUL_2558|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_6791|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0106|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0802|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_02461|M.leprae_Br4923 --------------------------------------------------
MAV_4681|M.avium_104 --------------------------------------------------
TH_2033|M.thermoresistible__bu MAENTIKWDKDADGIVTLTLDDPTGSANVMNEHYQESMRKAVDRLEAEKD
Rv1715|M.tuberculosis_H37Rv --------------------------------------------------
Mb1742|M.bovis_AF2122/97 --------------------------------------------------
MAB_4489c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1322|M.marinum_M --------------------------------------------------
MUL_2558|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_6791|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0106|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0802|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_02461|M.leprae_Br4923 --------------------------------------------------
MAV_4681|M.avium_104 --------------------------------------------------
TH_2033|M.thermoresistible__bu SITGVVITSAKKTFFAGGDLNGMMTIGPDDAQQAFDMVEAVKRDLRRLET
Rv1715|M.tuberculosis_H37Rv --------------------------------------------------
Mb1742|M.bovis_AF2122/97 --------------------------------------------------
MAB_4489c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1322|M.marinum_M --------------------------------------------------
MUL_2558|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_6791|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0106|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0802|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_02461|M.leprae_Br4923 --------------------------------------------------
MAV_4681|M.avium_104 --------------------------------------------------
TH_2033|M.thermoresistible__bu LGVPVVAAINGAALGGGLEIALACHRRIAADVKGSVIGLPEVTLGLLPGG
Rv1715|M.tuberculosis_H37Rv --------------------------------------------------
Mb1742|M.bovis_AF2122/97 --------------------------------------------------
MAB_4489c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1322|M.marinum_M --------------------------------------------------
MUL_2558|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_6791|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0106|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0802|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_02461|M.leprae_Br4923 --------------------------------------------------
MAV_4681|M.avium_104 --------------------------------------------------
TH_2033|M.thermoresistible__bu GGVTRTVRMFGIQKAFMEILSQGTRFRPAKAKEVGLVDELVDSVDELVPR
Rv1715|M.tuberculosis_H37Rv --------------------------------------------------
Mb1742|M.bovis_AF2122/97 --------------------------------------------------
MAB_4489c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1322|M.marinum_M --------------------------------------------------
MUL_2558|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_6791|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0106|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0802|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_02461|M.leprae_Br4923 --------------------------------------------------
MAV_4681|M.avium_104 --------------------------------------------------
TH_2033|M.thermoresistible__bu AKAWIKENPDAHTQPWDAKGYKMPGGTPSSPGLASILPSFPALLKKQLKG
Rv1715|M.tuberculosis_H37Rv --------------------------------------------------
Mb1742|M.bovis_AF2122/97 --------------------------------------------------
MAB_4489c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1322|M.marinum_M --------------------------------------------------
MUL_2558|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_6791|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0106|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0802|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_02461|M.leprae_Br4923 --------------------------------------------------
MAV_4681|M.avium_104 --------------------------------------------------
TH_2033|M.thermoresistible__bu APMPAPRAILNAAVEGAQVDFDSASRIESRYFTQLVTGQTAKNMIQAFFF
Rv1715|M.tuberculosis_H37Rv -------------MLTSHGFSRAAVVGAGLMGRRIAGVLASAGLDVAITD
Mb1742|M.bovis_AF2122/97 -------------MLTSHGFSRAAVVGAGLMGRRIAGVLASAGLDVAITD
MAB_4489c|M.abscessus_ATCC_199 ---------------MTNDIRPVCVVGGGRMGAGISQVFALAGFTVFVYE
MMAR_1322|M.marinum_M ------MTGAYTYTFADIRSRPVAVIGAGTLGRRIALMFASRGGTVRIYA
MUL_2558|M.ulcerans_Agy99 ------MTGAYTYTFADIRSRPVAVIGAGTLGRRIALMFASRGGTVRIYA
MSMEG_6791|M.smegmatis_MC2_155 ----------------MRKINTVTVVGAGYMGGGIAQVLALNGFKVQIAD
Mflv_0106|M.gilvum_PYR-GCK -----------------MSIERVGVVGAGQMGSGIAEVAAKAGANVVVYE
Mvan_0802|M.vanbaalenii_PYR-1 -----------------MSIERVGVVGAGQMGSGIAEVAAKAGANVVVYE
MLBr_02461|M.leprae_Br4923 --------------MSDAAIQRVGVVGAGQMGSGIAEVAVRAGVGVTVFE
MAV_4681|M.avium_104 ----MHDKTIGGETVSDAAIQRVGVVGAGQMGSGIAEVSVRADVDVTVFE
TH_2033|M.thermoresistible__bu DLQTINSGGSRPQGIEKTLPKKVGVLGAGMMGAGIAYVTAKAGFEVVLKD
. *:*.* :* *: : . . * :
Rv1715|M.tuberculosis_H37Rv TNAEILHAAAVEAARVAGAGRGSVAAAA--------------DLAAAIPD
Mb1742|M.bovis_AF2122/97 TNAEILHAAAVEAARVAGAGRGSVAAAA--------------DLAAAIPD
MAB_4489c|M.abscessus_ATCC_199 VNEQVRTSVVDRVRESLACAELDVAVADR--------IIITDDLSAAVAD
MMAR_1322|M.marinum_M RRAEQRAQATQYVADNLPKLLQDRGFGEV------GSVTATDCLATALEG
MUL_2558|M.ulcerans_Agy99 RRAEQRAQATQYVADNLPKLLQDRGFGEV------GSVTATDCLATALEG
MSMEG_6791|M.smegmatis_MC2_155 VNAEATQEALKRLDREAREFEQQGLFGPGSADTIMSNLTASDSLDDAVSD
Mflv_0106|M.gilvum_PYR-GCK PSDALVNAGRERITASLERAKAKGKLAADDFEI-TLARLTFTTDLADLAD
Mvan_0802|M.vanbaalenii_PYR-1 PSEALVAAGRDRITASLERAKAKGKLAADDFDA-TVARLTFTTNLGDLAD
MLBr_02461|M.leprae_Br4923 TTDTLVTAGRNRIVKSLERGVSAGKVTERERDS-ALGHLVFTTDLNDLAD
MAV_4681|M.avium_104 TTEALITAGRNRIVKSLERGVSAGKVTERERDR-ALSKLTFTTDLKDLAD
TH_2033|M.thermoresistible__bu VTLEAAQKGKGYSEKLEAKALERGRTTPEKSKA-LLDRIKPTADVADFKG
. .
Rv1715|M.tuberculosis_H37Rv ADLVIEAVVENLAVKQELFERL-ATLAP-DAVLATNTSVLPIGAVTERVE
Mb1742|M.bovis_AF2122/97 ADLVIEAVVENLAVKQELFERL-ATLAP-DAVLATNTSVLPIGAVTERVE
MAB_4489c|M.abscessus_ATCC_199 ACFVTEAVGEKLELKREVLAGI-AAAAPHDAILASNTSVIPISEIAAGSP
MMAR_1322|M.marinum_M AWLAVESVPEKLEIKTALWGQI-DQAAPPDTIFATNSSSFPSRLMADNVR
MUL_2558|M.ulcerans_Agy99 AWLAVESVPEKLEIKTALWGQI-DQAAPPDTIFATNSSSFPSRLMADNVR
MSMEG_6791|M.smegmatis_MC2_155 VDYVMEAVFEDVDVKKEVLARV-CASARPDTIIGTNTSTIPVKVLVDAVT
Mflv_0106|M.gilvum_PYR-GCK RQLVIEAIIEDEAVKAKVFAQLDEVVTDPDAVLASNTSSIPIMKIAAATK
Mvan_0802|M.vanbaalenii_PYR-1 RQLVIEAIVENEAVKAKVFAELDAVVTDPDAVLASNTSSIPIMKIAAATK
MLBr_02461|M.leprae_Br4923 RQLVIEAIVEDEAVKTKVFAELDRVITDPDAVLASNTSSIPIMKIAAATK
MAV_4681|M.avium_104 RQLVIEAIIEDDAVKAQVFAELDRVVTDPDAVLASNTSSIPIMKLAAATK
TH_2033|M.thermoresistible__bu VDFVIEAVFENTELKHKVFAEIEDIV-DPKAILGSNTSTLPITGLAAGVK
. *:: *. :* : : .:::.:*:* :* :.
Rv1715|M.tuberculosis_H37Rv DGSRVIGTHFWNPPDLIPVVEVVPSARTAPDTADRVVALLTQVGKLPVRV
Mb1742|M.bovis_AF2122/97 DGSRVIGTHFWNPPDLIPVVEVVPSARTAPDTADRVVALLTQVGKLPVRV
MAB_4489c|M.abscessus_ATCC_199 AIDRIVGTHWWNPAPLIPLVEVVQATGTSVETVSRTMKLLAAIGKKPMHV
MMAR_1322|M.marinum_M DKTRLCNTHFYMPPQFN-ALDLMSDGETDRGLLDTLLTVLPEFGVHPFEA
MUL_2558|M.ulcerans_Agy99 DKTRLCNTHFYMPPQFN-ALDLMSDGETDRGLLDTLLTVLPEFGVHPFEA
MSMEG_6791|M.smegmatis_MC2_155 HPERFLTVHFSNPAPFIPGVELVAGEATTQEVIDSVKDLLARAGREGAQV
Mflv_0106|M.gilvum_PYR-GCK NPSRVLGLHFFNPVPVLPLVEVINTLVTSEEAVARVEQFASEVLGKKVVR
Mvan_0802|M.vanbaalenii_PYR-1 NPSRVLGLHFFNPVPVLPLVELVNTLVTSDDAIARVERFAGEVLGKKVVR
MLBr_02461|M.leprae_Br4923 NPQRVLGLHFFNPVPVLPLVELVSTLVTDEAAATRAEEFASGVLGKQVVR
MAV_4681|M.avium_104 NPQRVLGLHFFNPVPVLPLVELVSTLVTDEAAAARTEEFASAVLGKQVVR
TH_2033|M.thermoresistible__bu RQEDFIGIHFFSPVDKMPLVEIIKGEKTSDEALARVFDYT-LAIGKTPIV
. *: * :::: *
Rv1715|M.tuberculosis_H37Rv GRDVPGFIGNRLQHALWREAIALVAEGVCDPKTVDLVVRNTIGLRLATLG
Mb1742|M.bovis_AF2122/97 GRDVPGFIGNRLQHAL----------------------------------
MAB_4489c|M.abscessus_ATCC_199 RKEVPGFVGNRLQHALWREAIALVQEGVCDATTVDDVVKNSFGLRLAVLG
MMAR_1322|M.marinum_M RRECTGFIFNRVWAAIKRESLAVVAEGVARPEDVDGMFKVNWRVP---AG
MUL_2558|M.ulcerans_Agy99 RRECTGFIFNRVWAAIKRESLAVVAEGVARPEDVDGMFKVNWRVP---AG
MSMEG_6791|M.smegmatis_MC2_155 A-DTPGMALNRLQYALLKEATLIVEEGVATKEDVDTIVRTTFGFRLGFFG
Mflv_0106|M.gilvum_PYR-GCK CGDRSGFIVNALLVPYLLSAIRMAEAGVATVEDIDTAVVAGLSHPMG---
Mvan_0802|M.vanbaalenii_PYR-1 CGDRSGFVVNALLVPYLLSAIRMAEAGVATVEDIDTAVVAGLSHPMG---
MLBr_02461|M.leprae_Br4923 CSDRSGFVVNALLVPYLLSAIRIVEAGFATVEDVDKAVLAGLSHPMG---
MAV_4681|M.avium_104 CSDRSGFVVNALLVPYLLSAIRMVEAGFATVEDVDKAVVAGLSHPMG---
TH_2033|M.thermoresistible__bu VNDSRGFYTSRVIGTFVNEALAMLGEGVEPASIEQAGGQAG--YPAP---
: *: . : .
Rv1715|M.tuberculosis_H37Rv PLENADYIGLDLTLAIHDAVIPSLNHDPH------PSPLLRELVAAGQLG
Mb1742|M.bovis_AF2122/97 --------------------------------------------------
MAB_4489c|M.abscessus_ATCC_199 PLENADLVGLDLTLDIHRVVVPALDCQGG------PNPVLVDKVARGMLG
MMAR_1322|M.marinum_M PFQLMDQVGLDVVLDIENHYADEFPYLPTN-----VRDLLRSYVDAGKLG
MUL_2558|M.ulcerans_Agy99 PFQLMDQVGLDVVLDIENHYADEFPYLPTN-----VRDLLRSYVDAGKLG
MSMEG_6791|M.smegmatis_MC2_155 PFAIADQAGLDVYVKGYHTLENAFG-ERMA-----TPKLLTDNVDAGRYG
Mflv_0106|M.gilvum_PYR-GCK PLRLSDLIGLDTMKLIADSMYDEYKDP-----NFAPPPLLQRMVEAGQLG
Mvan_0802|M.vanbaalenii_PYR-1 PLRLSDLIGLDTMKLIADSMYDEYKDP-----HYAPPPLLQRMVEAGQLG
MLBr_02461|M.leprae_Br4923 PLRLSDLVGLDTLKLIADKMFEEFKEP-----HYGPPPLLLRMVEAGLLG
MAV_4681|M.avium_104 PLRLSDLVGLDTLKLIADKMFEEFKEP-----QYGAPPLLLRMVEAGRLG
TH_2033|M.thermoresistible__bu PLQLSDELNLELMQKIATETRKGVEAAGGTYEPHPAEAVVNTMIELGRPG
Rv1715|M.tuberculosis_H37Rv ARTGHGFL----DWPAGAREATTARLAQHIAAQLQANEKGRGT-------
Mb1742|M.bovis_AF2122/97 --------------------------------------------------
MAB_4489c|M.abscessus_ATCC_199 CKTGEGFR----RWSPADIAEMNNRLEGHLVRLLHPRNTGPVG-------
MMAR_1322|M.marinum_M VKTGEGFY----TYPNSSDT------------------------------
MUL_2558|M.ulcerans_Agy99 VKTGEGFY----TYPNSSDT------------------------------
MSMEG_6791|M.smegmatis_MC2_155 TKNGKGWTGDFDDETKAAVIAYRNKAYSRMGELLRELGPAPKGA------
Mflv_0106|M.gilvum_PYR-GCK KKSGKGFYSY----------------------------------------
Mvan_0802|M.vanbaalenii_PYR-1 KKSGKGFYTY----------------------------------------
MLBr_02461|M.leprae_Br4923 KKSGQGFYTY----------------------------------------
MAV_4681|M.avium_104 KKTGQGFYTY----------------------------------------
TH_2033|M.thermoresistible__bu RAKGAGFYEYADGKRAGLWPGLREAFKSGSSTPPLQDMIDRMLFAEALET
Rv1715|M.tuberculosis_H37Rv --------------------------------------------------
Mb1742|M.bovis_AF2122/97 --------------------------------------------------
MAB_4489c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1322|M.marinum_M --------------------------------------------------
MUL_2558|M.ulcerans_Agy99 --------------------------------------------------
MSMEG_6791|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0106|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0802|M.vanbaalenii_PYR-1 --------------------------------------------------
MLBr_02461|M.leprae_Br4923 --------------------------------------------------
MAV_4681|M.avium_104 --------------------------------------------------
TH_2033|M.thermoresistible__bu QKCLDEGVITSTADANIGSIMGIGFPPWTGGTAQFIVGYQGELGTGKEAF
Rv1715|M.tuberculosis_H37Rv ----------------------
Mb1742|M.bovis_AF2122/97 ----------------------
MAB_4489c|M.abscessus_ATCC_199 ----------------------
MMAR_1322|M.marinum_M ----------------------
MUL_2558|M.ulcerans_Agy99 ----------------------
MSMEG_6791|M.smegmatis_MC2_155 ----------------------
Mflv_0106|M.gilvum_PYR-GCK ----------------------
Mvan_0802|M.vanbaalenii_PYR-1 ----------------------
MLBr_02461|M.leprae_Br4923 ----------------------
MAV_4681|M.avium_104 ----------------------
TH_2033|M.thermoresistible__bu VARAKQLAERYGDRFNPPESLL