For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MHKPTDPTEMTALELVAAYTSGELSPVEATAAVLSRIAQHDSAINAYCLVDEEAALAAATESEERWRGHY TRGLLDGVPISIKDVFLTRNWPTLRGSGLIDPDGPWDVDSPAVQRIRADGMVMVGKTTTPEFAWKGVTDS RQRGITHNPADPSRTAGGSSGGSAAAVAAGMAPMSIGTDGGGSVRIPASFCGVVGFKPTHGRIPMYPISP FGPLAHGGPITRTVEDAALLMDIISLPDYRDPTSLSPTLTTYRSQVYRDMTGLDIAYSPTLGYVDVDPEV ADLVENAVNALNDAGLPVTHADPGFSDPIDSFETLWAAGAAASLNNYGALDDVGTDIDPGLAQMWDTGAG ASATEYLAARNTCVQLGVTMGAFHELHNVLITPTVPIPAFPLGADVPPNSGMRWWAQWTPFTYPFNMTQQ PALSIPVGNTAQGLPVGMQIVGPRHSDDLVLAVGHFAERVLAG
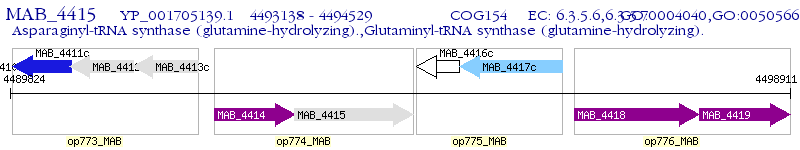
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4415 | - | - | 100% (463) | putative amidase |
| M. abscessus ATCC 19977 | MAB_4643c | - | 2e-59 | 36.12% (443) | amidase family protein |
| M. abscessus ATCC 19977 | MAB_3341c | gatA | 1e-52 | 32.85% (481) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3409 | amiD | 3e-51 | 31.15% (459) | amidase AmiD |
| M. gilvum PYR-GCK | Mflv_4246 | gatA | 2e-47 | 32.51% (483) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. tuberculosis H37Rv | Rv3375 | amiD | 3e-51 | 31.15% (459) | amidase AmiD |
| M. leprae Br4923 | MLBr_01702 | gatA | 5e-45 | 30.54% (478) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. marinum M | MMAR_3565 | amiC_1 | 1e-47 | 31.29% (473) | amidase AmiC_1 |
| M. avium 104 | MAV_3859 | gatA | 3e-48 | 32.08% (477) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
| M. smegmatis MC2 155 | MSMEG_2497 | - | 1e-138 | 54.50% (444) | putative amidase |
| M. thermoresistible (build 8) | TH_2639 | - | 1e-49 | 32.30% (483) | PUTATIVE glutamyl-tRNA (Gln) amidotransferase (subunit A), GatA |
| M. ulcerans Agy99 | MUL_2798 | amiC_1 | 6e-48 | 31.42% (471) | amidase AmiC_1 |
| M. vanbaalenii PYR-1 | Mvan_2115 | gatA | 3e-47 | 31.36% (491) | aspartyl/glutamyl-tRNA amidotransferase subunit A |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3409|M.bovis_AF2122/97 MTDADSAVPPRLDEDAISKLELTEVADLIRTRQLTSAEVTESTLRRIERL
Rv3375|M.tuberculosis_H37Rv MTDADSAVPPRLDEDAISKLELTEVADLIRTRQLTSAEVTESTLRRIERL
MAB_4415|M.abscessus_ATCC_1997 ----------MHKPTDPTEMTALELVAAYTSGELSPVEATAAVLSRIAQH
MSMEG_2497|M.smegmatis_MC2_155 -----------------MIPTSTDLVEAYRRKEMSPVEATQAALDAIDSY
Mflv_4246|M.gilvum_PYR-GCK -------------MSELIKLDAATLADKISSKEVSSAEVTQAFLDQIAAT
Mvan_2115|M.vanbaalenii_PYR-1 -------------MTDLIKLDAATLAAKIAAREVSATEVTQACLDQIAAT
MLBr_01702|M.leprae_Br4923 ---------MIGSLHEIIRLDASTLAAKIVAKELSSVEITQACLDQIEAT
MAV_3859|M.avium_104 -------------MNEIIRSDAATLAARIAAKELSSVEATQACLDQIAAT
TH_2639|M.thermoresistible__bu -------MSQGKSTDDLIRLDAATLGAKIAAKEISSTELTQACLDRIAAT
MMAR_3565|M.marinum_M ---------MSTLAEQTRWMDATDQAALVAAGEASPSELLEAAIERIERI
MUL_2798|M.ulcerans_Agy99 ---------MSTLAEQTRWMDATDQAALVVAGEASPSELLEAAIERIERI
: :. * : : *
Mb3409|M.bovis_AF2122/97 DPQLKSYAFVMPETALAAARAADA-----DIARGHYEGVLHGVPIGVKDL
Rv3375|M.tuberculosis_H37Rv DPQLKSYAFVMPETALAAARAADA-----DIARGHYEGVLHGVPIGVKDL
MAB_4415|M.abscessus_ATCC_1997 DSAINAYCLVDEEAALAAATESEE-----RWRGHYTRGLLDGVPISIKDV
MSMEG_2497|M.smegmatis_MC2_155 DKSVNAFVLVDHEGALAAAKESEA-----RWFSGEQLGPADGVPTSIKDA
Mflv_4246|M.gilvum_PYR-GCK DGDYHAFLHVGAEQALAAAQTVDR----AVAAGEHLPSPLAGVPLALKDV
Mvan_2115|M.vanbaalenii_PYR-1 DAEYHAFLHVAGDQALAAAATVDKAIHEATAAGERLPSPLAGVPLALKDV
MLBr_01702|M.leprae_Br4923 DDTYRAFLHVAAEKALSAAAAVDK----AVAAGGQLSSTLAGVPLALKDV
MAV_3859|M.avium_104 DERYHAFLHVAADEALGAAAVVDE----AVAAGERLPSPLAGVPLALKDV
TH_2639|M.thermoresistible__bu DDRYNAFLHVAANKALSAAARVDA----AVAAGEELPSPLAGVPLALKDV
MMAR_3565|M.marinum_M NPALNAVVIRWFDHARETASADLP------------GGPFRGVPFLIKDL
MUL_2798|M.ulcerans_Agy99 NPALNAVVIRWFDHARETASADLP------------GGPFRGVPFLIKDL
: .: : * :* . *** :**
Mb3409|M.bovis_AF2122/97 CYTVDAPTAAGT--TIFRD-FRPAYDATVVARLRAAGAVIIGKLAMTEGA
Rv3375|M.tuberculosis_H37Rv CYTVDAPTAAGT--TIFRD-FRPAYDATVVARLRAAGAVIIGKLAMTEGA
MAB_4415|M.abscessus_ATCC_1997 FLTRNWPTLRGS--GLIDPDGPWDVDSPAVQRIRADGMVMVGKTTTPEFA
MSMEG_2497|M.smegmatis_MC2_155 LWTRGWPTLRGS--TLIDEAGPWDEDAPSVARLREAGAVIIGKTTTPEYS
Mflv_4246|M.gilvum_PYR-GCK FTTTDMPTTCGS--KVLEG-WTSPYDATVTAKLRSAGIPILGKTNMDEFA
Mvan_2115|M.vanbaalenii_PYR-1 FTTTDMPTTCGS--KILEG-WTSPYDATVTAKLRAAGIPILGKTNMDEFA
MLBr_01702|M.leprae_Br4923 FTTVDMPTTCGS--KILQG-WHSPYDATVTTRLRAAGIPILGKTNMDEFA
MAV_3859|M.avium_104 FTTVDMPTTCGS--KILQG-WRSPYDATVTTKLRAAGIPILGKTNMDEFA
TH_2639|M.thermoresistible__bu FTTIDMPTTCGS--KILDG-WQAPYDATVTVALRAAGIPILGKTNLDEFA
MMAR_3565|M.marinum_M YALYAGQPISNGNLAFKQAHVIADADTTLVGRYRAAGLVIAGRTNSPELG
MUL_2798|M.ulcerans_Agy99 YALYAGQPISNGNLAFKQAHVIADADTTLVGRYRAAGLVIAGRTNSPELG
. . . *:. . * * : *: * .
Mb3409|M.bovis_AF2122/97 YLGYHPSLP--TPVNPWDPTAWAGVSSSGCGVATAAGLCFGSIGSDTGGS
Rv3375|M.tuberculosis_H37Rv YLGYHPSLP--TPVNPWDPTAWAGVSSSGCGVATAAGLCFGSIGSDTGGS
MAB_4415|M.abscessus_ATCC_1997 WKGVTDSRQRGITHNPADPSRTAGGSSGGSAAAVAAGMAPMSIGTDGGGS
MSMEG_2497|M.smegmatis_MC2_155 WKGVTDSYRYGATVNPWDTTKTAGGSSGGSAAAVGLGMGPWSVGTDGGGS
Mflv_4246|M.gilvum_PYR-GCK MGSSTENSAYGPTRNPWDTERVPGGSGGGSAAALAAFQAPLAIGTDTGGS
Mvan_2115|M.vanbaalenii_PYR-1 MGSSTENSAYGPTRNPWNVDCVPGGSGGGSAAALAAFQAPLAIGSDTGGS
MLBr_01702|M.leprae_Br4923 MGSSTENSAYGPTRNPWNVDRVPGGSGGGSAAALAAFQAPLAIGSDTGGS
MAV_3859|M.avium_104 MGSSTENSAYGPTRNPWNVERVPGGSGGGSAAALAAFQAPLAIGSDTGGS
TH_2639|M.thermoresistible__bu MGSSTENSAFGPTRNPWDVNRVPGGSGGGSAAALAAFQAPLAIGSDTGGS
MMAR_3565|M.marinum_M SLPTTEPVAWGPTRNPWDTSRTPGGSSGGSAAAVASGMVPFAHASDGGGS
MUL_2798|M.ulcerans_Agy99 SLPTTEPVAWGPTRNPWDTSRTPGGSSGGSAAAVASGMVPFAHASDGGGS
. ** : .* *..*...* . : .:* ***
Mb3409|M.bovis_AF2122/97 IRFPTSMCGVTGIKPTWGRVSRHGVVELAASYDHVGPITRSAHDAAVLLS
Rv3375|M.tuberculosis_H37Rv IRFPTSMCGVTGIKPTWGRVSRHGVVELAASYDHVGPITRSAHDAAVLLS
MAB_4415|M.abscessus_ATCC_1997 VRIPASFCGVVGFKPTHGRIPMYPISPFG-PLAHGGPITRTVEDAALLMD
MSMEG_2497|M.smegmatis_MC2_155 VRIPGGFTGTVALKPTYGLVPLYPMSAFG-SLAHVGPMTRSVRDCAALLD
Mflv_4246|M.gilvum_PYR-GCK IRQPAALTATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHQ
Mvan_2115|M.vanbaalenii_PYR-1 IRQPAALTATVGVKPTYGTVSRYGLIACASSLDQGGPCARTVLDTALLHQ
MLBr_01702|M.leprae_Br4923 IRQPAALTATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHA
MAV_3859|M.avium_104 IRQPAALTATVGVKPTYGTVSRYGLVACASSLDQGGPCARTVLDTALLHQ
TH_2639|M.thermoresistible__bu IRQPAALTATVGVKPTYGTVSRYGLVACASSLDQGGPCGRTVLDTALLHQ
MMAR_3565|M.marinum_M IRIPASCCGLVGLKPSQGRITLGPLREES-GMSVEHCLSRTVRDTAALLD
MUL_2798|M.ulcerans_Agy99 IRIPASCCGLVGLKPSQGRITLGPLREES-GMSVEHCLSRTVRDTAALLD
:* * . . ...**: * :. : . *:. * * *
Mb3409|M.bovis_AF2122/97 VIAGSDIHDPSCSAEPVPDYAADLALTRIP-----RVGVDWSQT--TSFD
Rv3375|M.tuberculosis_H37Rv VIAGSDIHDPSCSAEPVPDYAADLALTRIP-----RVGVDWSQT--TSFD
MAB_4415|M.abscessus_ATCC_1997 IISLPDYRDPTSLSPTLTTYRSQVYRDMTG----LDIAYSPTLG-YVDVD
MSMEG_2497|M.smegmatis_MC2_155 VISRFDGRDGTAMPTPTSSFLDGLDAGVAG----LRVAYSPNLG-YVHND
Mflv_4246|M.gilvum_PYR-GCK VIAGHDPLDSTSVEAPVPDVVAAARTGAGGDLTGVRIGVVKQLRSGEGYQ
Mvan_2115|M.vanbaalenii_PYR-1 VIAGHDPRDSTSVNAAVPDVVGAARAGARGDLKGVRIGVVKQLRSGEGYQ
MLBr_01702|M.leprae_Br4923 VIAGHDARDSTSVEAAVPDIVGAAKAGESGDLHGVRVGVVRQLR-GEGYQ
MAV_3859|M.avium_104 VIAGHDIRDSTSVDAPVPDVVGAARAGAAGDLKGVRVGVVKQLR-GEGYQ
TH_2639|M.thermoresistible__bu VIAGHDAKDSTSLTAPVPDVVAAARAGASGDLKGVRVGVVRQLR-GEGIQ
MMAR_3565|M.marinum_M ATRGPGVGDTVIAPPPIRPYTEELGADPGR----LRIGILDHHPRGGPVD
MUL_2798|M.ulcerans_Agy99 ATRGPGVGDTVIAPPPIRPYTEELGADPGR----LRIGILDHHPRGGPVD
. * . :. :
Mb3409|M.bovis_AF2122/97 EDTTAMLADVVKTLDDIGWPVIDVKLPALAPM--VAAFGKMRAVETAIAH
Rv3375|M.tuberculosis_H37Rv EDTTAMLADVVKTLDDIGWPVIDVKLPALAPM--VAAFGKMRAVETAIAH
MAB_4415|M.abscessus_ATCC_1997 PEVADLVENAVNALNDAGLPVTHAD-PGFSDP--IDSFETLWAAGAAASL
MSMEG_2497|M.smegmatis_MC2_155 PEVDAAVRAAVDVIAAAGAEVEEVD-PGFADP--VEAFHVLWFSGEAHVL
Mflv_4246|M.gilvum_PYR-GCK AGVLASFNAAVDQLTALGAEVTEVDCPHFDYS--LPAYYLILPSEVSSNL
Mvan_2115|M.vanbaalenii_PYR-1 PGVLASFTAAVEQLTALGAEVSEVDCPHFDHS--LAAYYLILPSEVSSNL
MLBr_01702|M.leprae_Br4923 SGVLASFQAAVEQLIALGATVSEVDCPHFDYA--LAAYYLILPSEVSSNL
MAV_3859|M.avium_104 PGVLASFEAAVEQLTALGAEVSEVDCPHFEYA--LAAYYLILPSEVSSNL
TH_2639|M.thermoresistible__bu PGVLASFDAAVEQLAALGAEVIEVDCPNIDHA--LAAYYLIMPSEVSSNL
MMAR_3565|M.marinum_M AEVTHATQAVGRLLESLGHHVDVAWPAALEDASFDSTFGALWSTNMGLAQ
MUL_2798|M.ulcerans_Agy99 AEVTHATQAVCRLLESLGHHVEVAWPAALEDASFDSTFGALWSTNMGLAQ
. . : * * . . : :: : .
Mb3409|M.bovis_AF2122/97 ADTYPARADEYG--------------------------PIMRAMIDAGHR
Rv3375|M.tuberculosis_H37Rv ADTYPARADEYG--------------------------PIMRAMIDAGHR
MAB_4415|M.abscessus_ATCC_1997 NNYGALDDVGTDIDPG------------------------LAQMWDTGAG
MSMEG_2497|M.smegmatis_MC2_155 RGYG--DAVDDRVDPG------------------------LRRTAAIGAT
Mflv_4246|M.gilvum_PYR-GCK AKFDGMRYGLRAGDDGTHSAEEVMALTRAAGFGPEVKRRIMIGAYALSAG
Mvan_2115|M.vanbaalenii_PYR-1 AKFDGMRYGLRVGDDGTTSAEEVMAMTRAAGFGAEVKRRIMIGTYALSAG
MLBr_01702|M.leprae_Br4923 ARFDAMRYGLRVGDDGSRSAEEVIALTRAAGFGPEVKRRIMIGAYALSAG
MAV_3859|M.avium_104 ARFDAMRYGLRIGDDGSHSAEEVMALTRAAGFGPEVKRRIMIGTYALSAG
TH_2639|M.thermoresistible__bu ARFDAMRYGLRVGDDGTRSAEEVMALTRAAGFGPEVKRRIMIGTYALSAG
MMAR_3565|M.marinum_M RRFEEQLGRPLGEDE---------------------FELVNRAQADFAKH
MUL_2798|M.ulcerans_Agy99 RRFEEQLGRPLGEDE---------------------FELVNRAQADFAKH
.
Mb3409|M.bovis_AF2122/97 LAAVEYQTLTERRLEFTRSLRRVFHD-VDILLMPSAGIASPTLETMRGLG
Rv3375|M.tuberculosis_H37Rv LAAVEYQTLTERRLEFTRSLRRVFHD-VDILLMPSAGIASPTLETMRGLG
MAB_4415|M.abscessus_ATCC_1997 ASATEYLAARNTCVQLGVTMGAFHEL-HNVLITPTVPIPAFPLGADVPPN
MSMEG_2497|M.smegmatis_MC2_155 FSAADYLDATAVRMDLGLRMGRFHQT-YDLLLTPTLPLPAFPAGQDVPDG
Mflv_4246|M.gilvum_PYR-GCK YYDAYYNQAQKVRTLIARDLDAAYQK-VDVLVSPATPTTAFRLGEKVDDP
Mvan_2115|M.vanbaalenii_PYR-1 YYDAYYNQAQKVRTLIARDLDEAYRS-VDVLVSPATPSTAFRLGEKVDDP
MLBr_01702|M.leprae_Br4923 YYDSYYSQAQKVRTLIARDLDKAYRS-VDVLVSPATPTTAFSLGEKADDP
MAV_3859|M.avium_104 YYDAYYNQAQKVRTLIARDLDAAYES-VDVVVSPATPTTAFGLGEKVDDP
TH_2639|M.thermoresistible__bu YYDAYYNQAQKVRTLVARDFDAAYEK-VDVLVSPTTPTTAFPLGEKVDHP
MMAR_3565|M.marinum_M FTSVDYALALSAIAAFRRAIQSWWHDGWDLLLTPTVASLPVKLGTIVSTP
MUL_2798|M.ulcerans_Agy99 FTSVDYALALSAIAAFRRAIQSWWHDGWDLLLTPTVASLPVKLGTIVSTP
* . : . :::: *: .
Mb3409|M.bovis_AF2122/97 QD----PELTARLAMPTAPFNVSGNPAICLPAGTTARG-TPLGVQFIGRE
Rv3375|M.tuberculosis_H37Rv QD----PELTARLAMPTAPFNVSGNPAICLPAGTTARG-TPLGVQFIGRE
MAB_4415|M.abscessus_ATCC_1997 SG----MRWWAQWTPFTYPFNMTQQPALSIPVGNTAQG-LPVGMQIVGPR
MSMEG_2497|M.smegmatis_MC2_155 WH----SPDWTSWTPYTYPFNLSQQPALTVPCGFTSAG-LPIGLQVVGPR
Mflv_4246|M.gilvum_PYR-GCK LS----MYLFDLCT---LPLNLAGHCGMSVPSGLSADDNLPVGLQIMAPA
Mvan_2115|M.vanbaalenii_PYR-1 LA----MYLFDLCT---LPLNLAGHCGMSVPSGLSADDNLPVGLQIMAPA
MLBr_01702|M.leprae_Br4923 LA----MYLFDLCT---LPLNLAGHCGMSVPSGLSSDDDLPVGLQIMAPA
MAV_3859|M.avium_104 LA----MYLFDLCT---LPLNLAGHCGMSVPSGLSPDDDLPVGLQIMAPA
TH_2639|M.thermoresistible__bu LA----MYLFDLCT---LPLNLAGHCGMSVPSGLSPDDNLPVGLQIMAPA
MMAR_3565|M.marinum_M EHPMAAMQRAGEFVPFTPPFNTSGQPAISLPLEWTPEG-LPLGVQLVAAY
MUL_2798|M.ulcerans_Agy99 EHPMAAMQRAGEFVPFTPPFNTSGQPAISLPLEWTPEG-LPLGVQLVAAY
. *:* : : .: :* :. . *:*:*.:.
Mb3409|M.bovis_AF2122/97 FDEHLLVRAGHAFQQVTG----YHRRRPPV--
Rv3375|M.tuberculosis_H37Rv FDEHLLVRAGHAFQQVTG----YHRRRPPV--
MAB_4415|M.abscessus_ATCC_1997 HSDDLVLAVGHFAE----------RVLAG---
MSMEG_2497|M.smegmatis_MC2_155 HSDSLVLRVGQAYQSATDWHLKTPKVLTGEGH
Mflv_4246|M.gilvum_PYR-GCK LADDRLYRVGAAYEAARG------PLPTAL--
Mvan_2115|M.vanbaalenii_PYR-1 LADDRLYRVGAAYEAARG------PLPTAL--
MLBr_01702|M.leprae_Br4923 LADDRLYRVGAAYEAARG------PLPSAI--
MAV_3859|M.avium_104 LADDRLYRVGAAYEAARG------PLRSAI--
TH_2639|M.thermoresistible__bu LADDRLYRVGAAYEAARG------PLPAAV--
MMAR_3565|M.marinum_M GREDVLLRIASQLEQAKP----WAQRTPDI--
MUL_2798|M.ulcerans_Agy99 GREDVLLRIASQLEQAKP----WAQRTPDI--
: : . : .