For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NVPLSVLDLSPVSEGADATTALRNSIDLAQHAEQWGYRRYWLAEHHFVAVASTVPALVIAHIAAATHRIR VGAGAVQIASTTPAAVVESFGILDALYPGRIDLGLGRTGHRIRDRLAGNEQKPEPAPAHEVDGLFIPRAP TEFGIKDNPKLRATAAALQQPGAHVADFDQQVADIQAFLDGSYVSPEGVALHIRPGEGADLPLWLFGSSK GESAQVAARRGLPFVANYHVTPWSTVEAVEAYRDAFQPSKALAEPYVIVSADAVAADDETTARHLTSTFG RWVHSIRSGHGAIPYPDPDVAQPLTEEERQHSEDRVATQFVGDAGQVAERLDTLARVTGADELVITSITH RHEDRRRSYELIAKEWGLI*
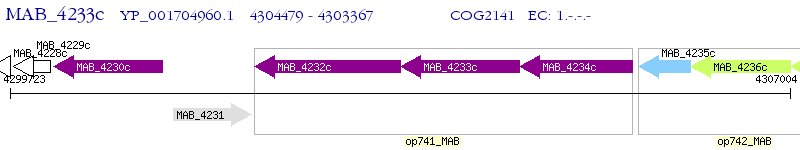
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4233c | - | - | 100% (370) | putative monooxygenase (luciferase-like) |
| M. abscessus ATCC 19977 | MAB_3639 | - | 5e-43 | 32.80% (372) | putative luciferase-like protein |
| M. abscessus ATCC 19977 | MAB_4628c | - | 2e-08 | 29.09% (110) | luciferase-like monooxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1971 | - | 1e-06 | 25.00% (272) | monooxygenase |
| M. gilvum PYR-GCK | Mflv_0215 | - | 1e-124 | 59.03% (371) | luciferase family protein |
| M. tuberculosis H37Rv | Rv0132c | fgd2 | 9e-07 | 37.23% (94) | putative f420-dependent glucose-6-phosphate dehydrogenase |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_0702 | - | 1e-118 | 58.38% (370) | monooxygenase-like hypothetical protein |
| M. avium 104 | MAV_2049 | - | 1e-129 | 61.41% (368) | hypothetical protein MAV_2049 |
| M. smegmatis MC2 155 | MSMEG_5029 | - | 2e-38 | 31.44% (388) | alkanal monooxygenase alpha chain |
| M. thermoresistible (build 8) | TH_4411 | - | 2e-38 | 30.35% (369) | PUTATIVE putative alkanal monooxygenase alpha chain |
| M. ulcerans Agy99 | MUL_4481 | - | 2e-39 | 32.63% (377) | monooxygenase |
| M. vanbaalenii PYR-1 | Mvan_4458 | - | 3e-39 | 31.19% (388) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0215|M.gilvum_PYR-GCK ----------MNVPLSVLDLAPICDGSDAATALRNT--------------
MMAR_0702|M.marinum_M -----------MIPLSILDLSPISAGSTAATALRNT--------------
MAV_2049|M.avium_104 ---------MTVTPLSILDLAPISAGCDAATALRNT--------------
MAB_4233c|M.abscessus_ATCC_199 ----------MNVPLSVLDLSPVSEGADATTALRNS--------------
MSMEG_5029|M.smegmatis_MC2_155 ------------MRLSVLDLVPVRTDQTTADALAAT--------------
Mvan_4458|M.vanbaalenii_PYR-1 -----------MMRLSVLDLVPVRSDQSTSDAIAAS--------------
MUL_4481|M.ulcerans_Agy99 ---------MGSVRLSVLDLVPVRTDQSTRDALAAT--------------
TH_4411|M.thermoresistible__bu ------------VPLSILDLVPISEGSTARDAIAAS--------------
Mb1971|M.bovis_AF2122/97 ------------MEIGIFLMPAHPPERTLYDATRWD--------------
Rv0132c|M.tuberculosis_H37Rv MTGISRRTFGLAAGFGAIGAGGLGGGCSTRSGPTPTPEPASRGVGVVLSH
:. : .
Mflv_0215|M.gilvum_PYR-GCK --------VDLARHAERWGYRRFWVAEHH----FVAVASTAPAVLIGQIA
MMAR_0702|M.marinum_M --------IELAQHAERWGYRRFWVAEHH----FVAVASASPAVLIGQIA
MAV_2049|M.avium_104 --------IDLARHAERWGYHRYWLAEHH----FVAVASSAPVTLIGQIA
MAB_4233c|M.abscessus_ATCC_199 --------IDLAQHAEQWGYRRYWLAEHH----FVAVASTVPALVIAHIA
MSMEG_5029|M.smegmatis_MC2_155 --------THLAQTADRLGYHRFWVAEHHN---MPAVAATSPPVLIAHLA
Mvan_4458|M.vanbaalenii_PYR-1 --------TLLAQTADRLGYTRYWVAEHHN---MPSVAATSPPVLIAHLA
MUL_4481|M.ulcerans_Agy99 --------VSLAQTAERLGFTRYWVAEHHN---MASVGATSPPVLIAYLA
TH_4411|M.thermoresistible__bu --------MNSARLADQLGYHRLWFAEHHN---TPNLAASATALLISQAA
Mb1971|M.bovis_AF2122/97 --------LDVIELADQLGYVEAWVGEHFT---VPWEPICAPDLLLAQAL
Rv0132c|M.tuberculosis_H37Rv EQFRTDRLVAHAQAAEQAGFRYVWASDHLQPWQDNEGHSMFPWLTLALVG
. *:: *: * .:* . :.
Mflv_0215|M.gilvum_PYR-GCK AATRTIRVGAAA-VQLGQTTAVAVVETFGMLDAFHPGRIDLGIGRSG-RR
MMAR_0702|M.marinum_M AATDRIRVGSAA-VQLSHTTAAAVVESFGMLDAFHPGRIDLGVGRSAQRR
MAV_2049|M.avium_104 AATSRIRVGTAA-VQIGHTTAVAVVENFGILDALYPGRIDMGLGRSGQRR
MAB_4233c|M.abscessus_ATCC_199 AATHRIRVGAGA-VQIASTTPAAVVESFGILDALYPGRIDLGLGRTGHRI
MSMEG_5029|M.smegmatis_MC2_155 AHTSQLRLGSGG-VMLPNHAPLAVAEQFALLEAAHPGRIDLGIGRAP---
Mvan_4458|M.vanbaalenii_PYR-1 AHTASMRVGSGG-VMLPNHAPLAVAEQFALLEAAHPGRIDLGIGRAP---
MUL_4481|M.ulcerans_Agy99 AQTTHLRLGSGG-VMLPNHAPLAVAEQFALLEAAAPGRIDLGIGRAP---
TH_4411|M.thermoresistible__bu TVTERIRVGAGG-VMLPNHAPLMVAEQYGTLANIHGDRIDLGLGRAP---
Mb1971|M.bovis_AF2122/97 LRTQQIKLAPGA-HLLPYHHPVELAHRVAYFDHLAQGRFMLGVGASG---
Rv0132c|M.tuberculosis_H37Rv NSTSSILFGTGVTCPIYRYHPATVAQAFASLAILNPGRVFLGLGTGER--
* : .... : . :.. . : .*. :*:*
Mflv_0215|M.gilvum_PYR-GCK ARPPRPDGQKRTPRPVPPWREIDGVVIPSPFDLSTLMRNPRLKARMSVLQ
MMAR_0702|M.marinum_M GVKPRGPAKSLAPQPAREWREIDGVVIPPPFDLRTLLSGGRVQATMAILQ
MAV_2049|M.avium_104 AEAAASSAAASPPE-ARPWRETDGLVIPTPFDQTALMKSERIRAQSSVLA
MAB_4233c|M.abscessus_ATCC_199 RDRLAGNEQK--PEPAPAH-EVDGLFIPRAPTEFGIKDNPKLRATAAALQ
MSMEG_5029|M.smegmatis_MC2_155 -----------------------GSDPVTSLALRG--AAGRDDRDIEAFP
Mvan_4458|M.vanbaalenii_PYR-1 -----------------------GSDPVTSLALRG--AAGRDERDIEAFP
MUL_4481|M.ulcerans_Agy99 -----------------------GSDPVTSYALRGGRSPAQQERDIESFP
TH_4411|M.thermoresistible__bu -----------------------GTDMMTARALSR------SSAEPQDFA
Mb1971|M.bovis_AF2122/97 ---------------------IPGDWALYDVDGKNGEHREMTREALEIML
Rv0132c|M.tuberculosis_H37Rv ------------------------LNEQAATDTFG-----NYRERHDRLI
:
Mflv_0215|M.gilvum_PYR-GCK QPGAVSPEFADQ-----VDDILALIAGRYE-IDGFAAHVVPGEGSALTPW
MMAR_0702|M.marinum_M QPEAVAPDFAEQ-----VGDILALLAGTYR-AEGFDVHAVPGENAALTPW
MAV_2049|M.avium_104 PPCAQPADFGEQ-----IADIEAMLDGSYA-VNGHRLQVVPGEHAELTPW
MAB_4233c|M.abscessus_ATCC_199 QPGAHVADFDQQ-----VADIQAFLDGSYVSPEGVALHIRPGEGADLPLW
MSMEG_5029|M.smegmatis_MC2_155 QYLDDVVALMGA-----RGVRVPLPRDLLR--DNYILKATPNATSVPQLW
Mvan_4458|M.vanbaalenii_PYR-1 QYLDDVVALMSS-----KGVRVPLPRDLMR--DNYILKATPAAVTEPRLW
MUL_4481|M.ulcerans_Agy99 DYLDDVVALMSA-----RGARVPLQ----S--GDYILKATSAALTQPRLW
TH_4411|M.thermoresistible__bu RNIYDLQGWFSD-----TGTAHSMP----------IVSAVSAGTRVP-IW
Mb1971|M.bovis_AF2122/97 RIWTEDEPWEHRGKYWNANGIAPMFEGLMR------RHIKPYQKPHPPIG
Rv0132c|M.tuberculosis_H37Rv EAIVLIRQLWSG-------ERISFTGHYFR---TDELKLYDTPAMPPPIF
.:
Mflv_0215|M.gilvum_PYR-GCK VFGSSK-GESARVAGARGLPFVASYHLTPATALDAVDVYRAHFR---PSA
MMAR_0702|M.marinum_M IFGSSK-GRSAQVAGALGLPFVASYHLTPATALEAIETYRDAFV---PSA
MAV_2049|M.avium_104 IFGSTR-GQSAEVAGARGLPFVASYHITPGTALDAIEAYRNAFR---PSA
MAB_4233c|M.abscessus_ATCC_199 LFGSSK-GESAQVAARRGLPFVANYHVTPWSTVEAVEAYRDAFQ---PSK
MSMEG_5029|M.smegmatis_MC2_155 LLGSS--MYSAHLAAAKGLPYVFAHHFSGHGTAEALAIYREEFQ---PSE
Mvan_4458|M.vanbaalenii_PYR-1 LLGSS--MYSAQLAAAKGLPYVFAHHFAGQGTEEALQYYRDNFR---PSD
MUL_4481|M.ulcerans_Agy99 LLGSS--MYSARLAAAKGLPYVFAHHFSGKGTEEALELYRSRFV---PSA
TH_4411|M.thermoresistible__bu VLGST--VNGAAIAAQLGLPFAVASHFAPDQLDAALRTYREMFSTEAPTA
Mb1971|M.bovis_AF2122/97 VTGFSAGSETLKLAGERGYIPMSLDLNTEYVATHWDAVEEGALR---SGR
Rv0132c|M.tuberculosis_H37Rv VAASGP--QSATLAGRYGDGWIAQARDIN--DAKLLAAFAAGAQ----AA
: . :*. *
Mflv_0215|M.gilvum_PYR-GCK TLTEPYVVVSADVVVAEDTATARHLASS--YGHWVH-AIRT--GDGAVPY
MMAR_0702|M.marinum_M ALQRPYVVVSADIVVAQDSATARRLASS--YGHWVY-SIRA--GGGAAPY
MAV_2049|M.avium_104 RLARPYVVVSADVVVAEDSATARHLASS--YGHWVY-SIRS--GQGAIPY
MAB_4233c|M.abscessus_ATCC_199 ALAEPYVIVSADAVAADDETTARHLTST--FGRWVH-SIRS--GHGAIPY
MSMEG_5029|M.smegmatis_MC2_155 AAPEPVTFLTVNAVVAETTEEAEALLLP--QLQMMA-RLRTGQPLNALDL
Mvan_4458|M.vanbaalenii_PYR-1 LTPEPVTFLTVNAAVAETHDDAMRLILP--NLQMMA-RLRTGQPMVPIDL
MUL_4481|M.ulcerans_Agy99 LAARPVTFLTVNAAVADTREEATALMMP--NLHMMA-RLRTGQPLGPVQL
TH_4411|M.thermoresistible__bu QIDRPRVMAGINVMVADTDEEARRQFTV--VEQMFL-DLKHNLRR-KIQP
Mb1971|M.bovis_AF2122/97 TPDRRDWRLVREVLVAETDEQAFRYAVDGTMGRAMR-EYVLPTFRMFGMT
Rv0132c|M.tuberculosis_H37Rv GRDPTTLGKRAELFAVVGDDKAAARAAD--LWRFTAG-----------AV
: .. * :
Mflv_0215|M.gilvum_PYR-GCK PDPDRVAPLPDEQAATVKDRTATQFVGDPDEVAHRLEALRRVAD-ADELV
MMAR_0702|M.marinum_M PDPDDCAALTDDQLAVVSDRLATQFVGNPDEVAERMLALQRVAG-ADELV
MAV_2049|M.avium_104 PDPDLCDPLTEEQRALVADRTATQFVGNPDEVADRLEVLRRVSG-ADELV
MAB_4233c|M.abscessus_ATCC_199 PDPDVAQPLTEEERQHSEDRVATQFVGDAGQVAERLDTLARVTG-ADELV
MSMEG_5029|M.smegmatis_MC2_155 VEDAQTTVLTPQGDAVVADGRRRAVVGSPTEAAEQVRALAEEFD-VDEVM
Mvan_4458|M.vanbaalenii_PYR-1 VEDAEAQVVSPRAQAVIDAGLRQAVVGSPAEAGDQLRALAEKFG-VDEVM
MUL_4481|M.ulcerans_Agy99 VEEAQDARLTPEQQRIVDSGLERAVVGSPEEAAEQVRALGQRFG-VDEVM
TH_4411|M.thermoresistible__bu PVDPDTLAAQGGGN---EPMLRIKAVGSPPTVKAQLDEFVARTG-ADELI
Mb1971|M.bovis_AF2122/97 KFYKHNPSVPDDEVTPEYLAENTFVVGSVQTVVDKLEATYDQVGGFGHLL
Rv0132c|M.tuberculosis_H37Rv DQPNPVEIQRAAESNPIEKVLANWAVGTDPGVHIGAVQAVLDAG-AVPFL
** . . .:
Mflv_0215|M.gilvum_PYR-GCK ITSVTHGHE--------DRLRSHRLLAERWGITGEG------
MMAR_0702|M.marinum_M VTSVTHGHQ--------DRLRSHELIARRWGLWS--------
MAV_2049|M.avium_104 ITSVTHRHA--------DRLRSHELIAKRWGLTG--------
MAB_4233c|M.abscessus_ATCC_199 ITSITHRHE--------DRRRSYELIAKEWGLI---------
MSMEG_5029|M.smegmatis_MC2_155 INPVASAHRGADPRTATARDTTLELLAKELF-----------
Mvan_4458|M.vanbaalenii_PYR-1 VHPVASAFRGVDPATAPAREATLELLAKEIF-----------
MUL_4481|M.ulcerans_Agy99 VHPVASAHRGTGPATAPARVATCELLAKELF-----------
TH_4411|M.thermoresistible__bu T--VTYAHD------PAVRDRSLELLAKAWF-----------
Mb1971|M.bovis_AF2122/97 ILGFDYSDN-PGPWKESLRLLAHEVMPRLNARLATKPATAVV
Rv0132c|M.tuberculosis_H37Rv HFPQDDPIT-------AIDFYRTNVLPELR------------
.::..