For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSHVAADARLEPAADDWTETPDVESGSKSEDDHTSDPAAGALPQLLDRAPRLIKRPVAYAVSQTDSMLKT LGRYVDLVVQSFAFLISDIVRLRHPWQDTVIQSWFILTVTVVPAILVSIPFGVIVAVQAGSIISQVGASS ISGAAGGLGVIQQGAPIVAALLLGGAAGSAVATDLGARSIREEVDAMRVMGVNPVQRLVTPRLAAILFVS PLLCIFIIFVGIFAGYLVNVGFQGGTPGSYLSSFAAFANVSDVTVAVLKTWLFGVVVILVACQRGLEAKG GARGVADAVNATVVIGVVTVMVLNTVITQIVAMFMPSKVG
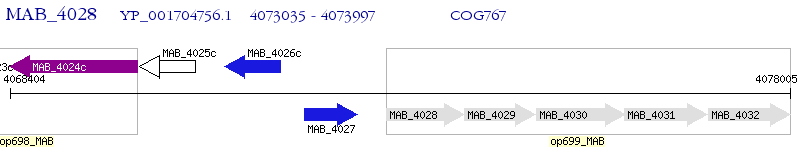
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4028 | - | - | 100% (320) | putative YrbE family protein |
| M. abscessus ATCC 19977 | MAB_4601c | - | 7e-62 | 44.98% (249) | putative YrbE family protein |
| M. abscessus ATCC 19977 | MAB_4518c | - | 3e-59 | 44.18% (249) | putative YrbE family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3531c | yrbE4A | 1e-48 | 42.27% (220) | integral membrane protein YrbE4a |
| M. gilvum PYR-GCK | Mflv_0012 | - | 2e-57 | 43.37% (249) | hypothetical protein Mflv_0012 |
| M. tuberculosis H37Rv | Rv3501c | yrbE4A | 1e-48 | 42.27% (220) | integral membrane protein YrbE4a |
| M. leprae Br4923 | MLBr_02587 | yrbE1A | 2e-47 | 36.05% (258) | hypothetical protein MLBr_02587 |
| M. marinum M | MMAR_0175 | yrbE6A | 4e-61 | 42.59% (270) | integral membrane protein YrbE6A |
| M. avium 104 | MAV_5054 | - | 3e-63 | 46.15% (234) | ABC transporter integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_1141 | - | 8e-59 | 43.78% (249) | ABC-transporter integral membrane protein |
| M. thermoresistible (build 8) | TH_2930 | - | 8e-59 | 43.78% (249) | PUTATIVE protein of unknown function DUF140 |
| M. ulcerans Agy99 | MUL_4921 | yrbE6A | 5e-61 | 42.22% (270) | integral membrane protein YrbE6A |
| M. vanbaalenii PYR-1 | Mvan_0957 | - | 6e-59 | 43.78% (249) | hypothetical protein Mvan_0957 |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_4028|M.abscessus_ATCC_1997 MSHVAADARLEPAADDWTETPDVESGSKSEDDHTSDPAAGALPQLLDRAP
MAV_5054|M.avium_104 --------------------------------------------------
Mflv_0012|M.gilvum_PYR-GCK --------MRLPGPRTRCGRRHRRSECTAAERTSGMTTADQPVRRDDSVH
Mvan_0957|M.vanbaalenii_PYR-1 -----------------------------------MSSDTK----GEGVG
TH_2930|M.thermoresistible__bu -----------------------------------MT-------RSDGVE
MSMEG_1141|M.smegmatis_MC2_155 ----------------------------MTTSPDRVSDGGAP---ERGTD
MMAR_0175|M.marinum_M -----------------------------------MATTFVN----DRLI
MUL_4921|M.ulcerans_Agy99 -----------------------------------MATTFVN----DRLI
Mb3531c|M.bovis_AF2122/97 --------------------------------------------------
Rv3501c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_02587|M.leprae_Br4923 -----------------------------------------------MTT
MAB_4028|M.abscessus_ATCC_1997 RLIKRPVAYAVSQTDSMLKTLGRYVDLVVQSFAFLISDIVRLRHPWQDTV
MAV_5054|M.avium_104 -----------------------------------MTDTLTLRLPFGEVI
Mflv_0012|M.gilvum_PYR-GCK AVQDWVGGYVRRHPLASLATVGDQFVLGVRTLQWFFVDLFTGRFQFQEFV
Mvan_0957|M.vanbaalenii_PYR-1 VITDWAGGYVKRHPLASLTTVGDQFVLGVRTVQYFLLDLFSGRFQWQEFI
TH_2930|M.thermoresistible__bu VIGEWAGGYIRRHPLAALGTVGEQFVLGVRTLQYLFVDLFTGRFQWWEFV
MSMEG_1141|M.smegmatis_MC2_155 AIEKWATGYVRRHPIASLTTVGDQFVLGVRTVQYLFTELFTGRFQWQEFI
MMAR_0175|M.marinum_M GLRRLSVTYLRQHPAASLATAGEQIVLGVAALRYLAIDLVTGRFPWTECI
MUL_4921|M.ulcerans_Agy99 GLRRLSVTYLRQHPAASLATAGEQIVLGVAALRYLAIDLVTGRFPWTECI
Mb3531c|M.bovis_AF2122/97 ---------MIQQLAVPARAVGGFFEMSMDTAR----AAFRRPFQFREFL
Rv3501c|M.tuberculosis_H37Rv ---------MIQQLAVPARAVGGFFEMSMDTAR----AAFRRPFQFREFL
MLBr_02587|M.leprae_Br4923 ETRSSLVEYAHTKLQTPLVLVGGFFRMLVLTGK----ALFRRPFQWREFI
. : : :
MAB_4028|M.abscessus_ATCC_1997 IQSWFILTVTVVPAILVSIPFGVIVAVQAGSIISQVGASSISGAAGGLGV
MAV_5054|M.avium_104 VQAWTLLKVTALPAVLMAIPFGAMVAVQLSGLVNEVGANSLVGSATGVAV
Mflv_0012|M.gilvum_PYR-GCK RQGAFMAGTAVLPTVLVALPIGVTLSIQFALLAGQVGATSLAGAASGLAV
Mvan_0957|M.vanbaalenii_PYR-1 RQGAFMAGTAVLPTVLVALPIGVTLSIQFALLAGQVGATSLAGAASGLAV
TH_2930|M.thermoresistible__bu RQAAFMAGTAVLPTVLVALPIGVTLSIQFALLAGQVGATSLAGAASGLAV
MSMEG_1141|M.smegmatis_MC2_155 RQGAFMASTAVVPTVLVALPIGVTLSIQFALLAGQVGATSLAGAASGLAV
MMAR_0175|M.marinum_M RQASFMAATAIMPTILVALPIGATLSIQFAMLAGQVGATSLAGAASGLTV
MUL_4921|M.ulcerans_Agy99 RQASFMAATAIMPTILVALPIGATLSIQFAMLAGQVGATSLAGAASGLTV
Mb3531c|M.bovis_AF2122/97 DQTWMVARVSLVPTLLVSIPFTVLVAFTLNILLREIGAADLSGAGTAFGT
Rv3501c|M.tuberculosis_H37Rv DQTWMVARVSLVPTLLVSIPFTVLVAFTLNILLREIGAADLSGAGTAFGT
MLBr_02587|M.leprae_Br4923 LQCWFIMRVAFLPTVMVSIPLTVLLIFTLNVLLAQFSAADLSGAGAAIGA
* : .: :*::::::*: . : . : :..* .: *:. .. .
MAB_4028|M.abscessus_ATCC_1997 IQQGAPIVAALLLGGAAGSAVATDLGARSIREEVDAMRVMGVNPVQRLVT
MAV_5054|M.avium_104 LRQGAPVTAGLLMGGAAASAIASDFGARAIREELDALKTLGIDPVRRLVV
Mflv_0012|M.gilvum_PYR-GCK IRQAASLTAAILMAAAVGSAITADLGSRKMREETDAMEVMGVSVIRRLVV
Mvan_0957|M.vanbaalenii_PYR-1 IRQAASLTAAILMAAAVGSAITADLGSRTMREETDAMEVMGVSVIRRLVV
TH_2930|M.thermoresistible__bu IRQAASLTAAILMAAAVGSAITADLGSRTMREETDAMEVMGVSVIRRLVV
MSMEG_1141|M.smegmatis_MC2_155 IRQAASLTAAVLMAAAVGSAITADLGSRKMREETDAMEVMGVSVIKRLVV
MMAR_0175|M.marinum_M VRQAASLVAAILLAAAVGSAITADLGVRTMREETDAMEVMGVSVVRNLVV
MUL_4921|M.ulcerans_Agy99 VRQAASLVAAILLAAAVGSAITADLGVRTMREETDAMEMMGVSVVRNLVV
Mb3531c|M.bovis_AF2122/97 ITQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMRVLGIDPIQRLVV
Rv3501c|M.tuberculosis_H37Rv ITQLGPVVTVLVVAGAGATAICADLGARTIREEIDAMRVLGIDPIQRLVV
MLBr_02587|M.leprae_Br4923 VTQLGPLTTVLVVAGAGSTSICADLGARTIREEIDAMEVLGIDPIHRLVV
: * ..:.: :::..* .::: :*:* * :*** **:. :*:. ::.**.
MAB_4028|M.abscessus_ATCC_1997 PRLAAILFVSPLLCIFIIFVGIFAGYLVNVGFQGGTPGSYLSSFAAFANV
MAV_5054|M.avium_104 PRFLGLLLITPSLVVIVIAMGVGAAFVIATVVNGVTPGSFWLSFGSFAKM
Mflv_0012|M.gilvum_PYR-GCK PRFAAAIMIGVALTGVVCFVGFLASYLFNVYFQNGAPGSFVATFASFATT
Mvan_0957|M.vanbaalenii_PYR-1 PRFAAAIMIGVALTGVVCFVGFLASYLFNVYFQNGAPGSFVATFASFATT
TH_2930|M.thermoresistible__bu PRFAAAIMIGVALTGVVCFVGFLASYLFNVYFQNGAPGSFVATFASFTTT
MSMEG_1141|M.smegmatis_MC2_155 PRFAAAIMIGVALTGVVCFVGFIASFLFNVYFQDGAPGSFVATFASFATT
MMAR_0175|M.marinum_M PRLLAAMVIGVALCGVVCFVGFLASYLFNVYVQGGSAGSFLATFSAFATT
MUL_4921|M.ulcerans_Agy99 PRLLAAMVIGVALCGVVCFVGFLASYLFNVYVQGGSAGSFLATFSAFATT
Mb3531c|M.bovis_AF2122/97 PRVLASTLVALLLNGLVCAIGLSGGYAFSVFLQGVNPGAFINGLTVLTGL
Rv3501c|M.tuberculosis_H37Rv PRVLASTLVALLLNGLVCAIGLSGGYAFSVFLQGVNPGAFINGLTVLTGL
MLBr_02587|M.leprae_Br4923 PRVLAATLVATLLNGLVITVGLVGGYLFGVYLQNVSGGAYLATLTTITGL
**. . .: * .: :*. ..: . . .:. *:: : ::
MAB_4028|M.abscessus_ATCC_1997 SDVTVAVLKTWLFGVVVILVACQRGLEAKGGARGVADAVNATVVIGVVTV
MAV_5054|M.avium_104 VDLWFTMGKGFIFAGIVAVISSQRGMEAKGGPRGVADAVNASVVLNVIFI
Mflv_0012|M.gilvum_PYR-GCK GDMIVALVKAVIFGAIVAIVSCQKGLSTQGGPTGVANSVNAAVVESILVL
Mvan_0957|M.vanbaalenii_PYR-1 GDMIVALVKAVIFGAIVAVVSCQKGLATQGGPTGVANSVNAAVVESILLL
TH_2930|M.thermoresistible__bu GDMLVALIKAVVFGAIVAVVSCQKGLSTKGGPTGVANSVNAAVVESILVL
MSMEG_1141|M.smegmatis_MC2_155 GDMIVALIKAVIFGAIVAVVSCQKGLSTVGGPTGVANSVNAAVVESILLL
MMAR_0175|M.marinum_M GDMILALLKAAVYGFIVTVVACQKGLSTRGGPAGVANSVNAAVVEAVLLL
MUL_4921|M.ulcerans_Agy99 GDMILALLKAAVYGFIVTVVACQKGLSTRGGPAGVANSVNAAVVEAVLLL
Mb3531c|M.bovis_AF2122/97 RELILAEIKALLFGVMAGLVGCYRGLTVKGGPKGVGNAVNETVVYAFICL
Rv3501c|M.tuberculosis_H37Rv RELILAEIKALLFGVMAGLVGCYRGLTVKGGPKGVGNAVNETVVYAFICL
MLBr_02587|M.leprae_Br4923 PEVVIATVKAATFGLIAGLVGCYRGLIVRGGSNGLGAAVNETVVLCVVAL
:: .: * :. :. ::.. :*: . **. *:. :** :** .: :
MAB_4028|M.abscessus_ATCC_1997 MVLNTVITQIVAMFMPSKVG
MAV_5054|M.avium_104 VVVNLAITQLQTMFFPMAVA
Mflv_0012|M.gilvum_PYR-GCK MVVNVAISQLYILMFPRVGL
Mvan_0957|M.vanbaalenii_PYR-1 MVVNVAISQLYIMMFPRVGL
TH_2930|M.thermoresistible__bu MIVNVGISQLYIMLFPRVGL
MSMEG_1141|M.smegmatis_MC2_155 MVVNVAISQLYIMLFPRVGL
MMAR_0175|M.marinum_M MIVNVVVSELYTVLFPRTGL
MUL_4921|M.ulcerans_Agy99 MIVNVVVSELYTVLFPRTGL
Mb3531c|M.bovis_AF2122/97 FVINVVMTAIGVRISAQ---
Rv3501c|M.tuberculosis_H37Rv FVINVVMTAIGVRISAQ---
MLBr_02587|M.leprae_Br4923 YAVNVVLTTIGVRFGTGH--
:* :: : : .