For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTSPDRVSDGGAPERGTDAIEKWATGYVRRHPIASLTTVGDQFVLGVRTVQYLFTELFTGRFQWQEFIR QGAFMASTAVVPTVLVALPIGVTLSIQFALLAGQVGATSLAGAASGLAVIRQAASLTAAVLMAAAVGSAI TADLGSRKMREETDAMEVMGVSVIKRLVVPRFAAAIMIGVALTGVVCFVGFIASFLFNVYFQDGAPGSFV ATFASFATTGDMIVALIKAVIFGAIVAVVSCQKGLSTVGGPTGVANSVNAAVVESILLLMVVNVAISQLY IMLFPRVGL
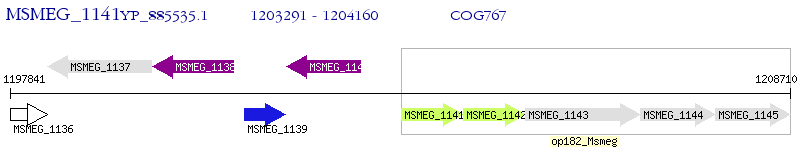
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1141 | - | - | 100% (289) | ABC-transporter integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_5902 | - | 1e-45 | 37.07% (232) | hypothetical protein MSMEG_5902 |
| M. smegmatis MC2 155 | MSMEG_0132 | - | 2e-45 | 35.71% (252) | hypothetical protein MSMEG_0132 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0173 | yrbE1A | 1e-46 | 38.34% (253) | integral membrane protein YRBE1A |
| M. gilvum PYR-GCK | Mflv_0012 | - | 1e-134 | 90.00% (270) | hypothetical protein Mflv_0012 |
| M. tuberculosis H37Rv | Rv0167 | yrbE1A | 1e-46 | 38.34% (253) | integral membrane protein YRBE1A |
| M. leprae Br4923 | MLBr_02587 | yrbE1A | 2e-44 | 39.19% (222) | hypothetical protein MLBr_02587 |
| M. abscessus ATCC 19977 | MAB_1012c | - | 1e-126 | 78.89% (289) | putative YrbE family protein |
| M. marinum M | MMAR_3862 | yrbE5A | 1e-129 | 82.01% (289) | YrbE family protein, YrbE5A |
| M. avium 104 | MAV_2390 | - | 1e-127 | 83.52% (273) | hypothetical protein MAV_2390 |
| M. thermoresistible (build 8) | TH_2930 | - | 1e-133 | 88.28% (273) | PUTATIVE protein of unknown function DUF140 |
| M. ulcerans Agy99 | MUL_3794 | yrbE5A | 1e-129 | 80.97% (289) | YrbE family protein, YrbE5A |
| M. vanbaalenii PYR-1 | Mvan_0957 | - | 1e-135 | 90.84% (273) | hypothetical protein Mvan_0957 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1141|M.smegmatis_MC2_155 -----------------------------MTTSPDRVSDGGAPERGTDAI
Mvan_0957|M.vanbaalenii_PYR-1 -------------------------------MSSDTKGEG------VGVI
Mflv_0012|M.gilvum_PYR-GCK MRLPGPRTRCGRRHRRSECTAAERTSGMTTADQPVRRDDS------VHAV
TH_2930|M.thermoresistible__bu ----------------------------------MTRSDG------VEVI
MMAR_3862|M.marinum_M -----------------------------MTTQPEPALLGSRNDDGVAAI
MUL_3794|M.ulcerans_Agy99 -----------------------------MTTQPEPALLGSRNDDGVAAI
MAV_2390|M.avium_104 -----------------------------MTATLTTPLDG---VDGVAAI
MAB_1012c|M.abscessus_ATCC_199 -----------------------------MTTQDERRATN----DGVAAI
Mb0173|M.bovis_AF2122/97 -----------------------------MTTST----------------
Rv0167|M.tuberculosis_H37Rv -----------------------------MTTST----------------
MLBr_02587|M.leprae_Br4923 -----------------------------MTTETRS--------------
MSMEG_1141|M.smegmatis_MC2_155 EKWATGYVRRHPIASLTTVGDQFVLGVRTVQYLFTELFTGRFQWQEFIRQ
Mvan_0957|M.vanbaalenii_PYR-1 TDWAGGYVKRHPLASLTTVGDQFVLGVRTVQYFLLDLFSGRFQWQEFIRQ
Mflv_0012|M.gilvum_PYR-GCK QDWVGGYVRRHPLASLATVGDQFVLGVRTLQWFFVDLFTGRFQFQEFVRQ
TH_2930|M.thermoresistible__bu GEWAGGYIRRHPLAALGTVGEQFVLGVRTLQYLFVDLFTGRFQWWEFVRQ
MMAR_3862|M.marinum_M GDWAAGYVRRHPLAALTTVGEQFVLGVRTIQYFFVDLVTGRFQWQEFVRQ
MUL_3794|M.ulcerans_Agy99 GDWAAGYIRRHPLAALTTVGEQFVLGVRTIQYFFVDLVTGRFQWQEFVRQ
MAV_2390|M.avium_104 RNWSVGYVKRHPVASLTTVGEQFVLGVRTIQYFFYDLITGRFQWQEFVRQ
MAB_1012c|M.abscessus_ATCC_199 QDWTTGYVKRHPLESLKTVGEQFMLGVRTIQWFFVDLFTGNFQWQEFVRQ
Mb0173|M.bovis_AF2122/97 --TLGGYVRDQLQTPLTLVGGFFRMCVLTGK----ALFRWPFQWREFILQ
Rv0167|M.tuberculosis_H37Rv --TLGGYVRDQLQTPLTLVGGFFRMCVLTGK----ALFRWPFQWREFILQ
MLBr_02587|M.leprae_Br4923 --SLVEYAHTKLQTPLVLVGGFFRMLVLTGK----ALFRRPFQWREFILQ
* : : .* ** * : * * : *. **: **: *
MSMEG_1141|M.smegmatis_MC2_155 GAFMASTAVVPTVLVALPIGVTLSIQFALLAGQVGATSLAGAASGLAVIR
Mvan_0957|M.vanbaalenii_PYR-1 GAFMAGTAVLPTVLVALPIGVTLSIQFALLAGQVGATSLAGAASGLAVIR
Mflv_0012|M.gilvum_PYR-GCK GAFMAGTAVLPTVLVALPIGVTLSIQFALLAGQVGATSLAGAASGLAVIR
TH_2930|M.thermoresistible__bu AAFMAGTAVLPTVLVALPIGVTLSIQFALLAGQVGATSLAGAASGLAVIR
MMAR_3862|M.marinum_M GAFMAGTAVLPTILVSLPISVTLSIQFALLAGQVGATSLAGAASGLAVIR
MUL_3794|M.ulcerans_Agy99 GAFMAGTAVLPTILVSLPISVTLSIQFALLAGQVGATSLAGAASGLAVIR
MAV_2390|M.avium_104 GAFMAGTAVLPTILVSLPISVTLSIQFALLAGQVGATSLAGAASGLAVIR
MAB_1012c|M.abscessus_ATCC_199 GAFQAGTAVVPVILVTLPIGVTLSIQFALLAGQVGATSLAGAASGIAIIR
Mb0173|M.bovis_AF2122/97 CWFIMRVGFLPTIMVSIPLTVLLIFTLNILLAQFGAADISGSGAAIGAVT
Rv0167|M.tuberculosis_H37Rv CWFIMRVGFLPTIMVSIPLTVLLIFTLNILLAQFGAADISGSGAAIGAVT
MLBr_02587|M.leprae_Br4923 CWFIMRVAFLPTVMVSIPLTVLLIFTLNVLLAQFSAADLSGAGAAIGAVT
* ...:*.::*::*: * * : : :* .*..*:.::*:.:.:. :
MSMEG_1141|M.smegmatis_MC2_155 QAASLTAAVLMAAAVGSAITADLGSRKMREETDAMEVMGVSVIKRLVVPR
Mvan_0957|M.vanbaalenii_PYR-1 QAASLTAAILMAAAVGSAITADLGSRTMREETDAMEVMGVSVIRRLVVPR
Mflv_0012|M.gilvum_PYR-GCK QAASLTAAILMAAAVGSAITADLGSRKMREETDAMEVMGVSVIRRLVVPR
TH_2930|M.thermoresistible__bu QAASLTAAILMAAAVGSAITADLGSRTMREETDAMEVMGVSVIRRLVVPR
MMAR_3862|M.marinum_M QGASLVAAVLMASAVGSAITADLGSRTMREETDAMEVMGVSVIRRLVVPR
MUL_3794|M.ulcerans_Agy99 QGASLVAAVLMASAVGSAITADLGSRTMREETDAMEVMGLSVIRRLVVPR
MAV_2390|M.avium_104 QGASLVAAVLMASAVGSAITADLGSRTMREETDAMEVMGVSVVRRLVVPR
MAB_1012c|M.abscessus_ATCC_199 QAASLVAALLMSAAVGSAITADLGSRTMREETDAMEVMGVSVIRRLVVPR
Mb0173|M.bovis_AF2122/97 QLGPLTTVLVVAGAGSTAICADLGARTIREEIDAMEVLGIDPIHRLVVPR
Rv0167|M.tuberculosis_H37Rv QLGPLTTVLVVAGAGSTAICADLGARTIREEIDAMEVLGIDPIHRLVVPR
MLBr_02587|M.leprae_Br4923 QLGPLTTVLVVAGAGSTSICADLGARTIREEIDAMEVLGIDPIHRLVVPR
* ..*.:.::::.* .::* ****:*.:*** *****:*:. ::******
MSMEG_1141|M.smegmatis_MC2_155 FAAAIMIGVALTGVVCFVGFIASFLFNVYFQDGAPGSFVATFASFATTGD
Mvan_0957|M.vanbaalenii_PYR-1 FAAAIMIGVALTGVVCFVGFLASYLFNVYFQNGAPGSFVATFASFATTGD
Mflv_0012|M.gilvum_PYR-GCK FAAAIMIGVALTGVVCFVGFLASYLFNVYFQNGAPGSFVATFASFATTGD
TH_2930|M.thermoresistible__bu FAAAIMIGVALTGVVCFVGFLASYLFNVYFQNGAPGSFVATFASFTTTGD
MMAR_3862|M.marinum_M FAAAIMIGVALTGVVCFVGFFASYMFNVYFQNGAPGSFVATFASFATTDD
MUL_3794|M.ulcerans_Agy99 FAAAIMIGVALTGVVCFVGFFASYMFNVYFQNGAPGSFVATFASFATTDD
MAV_2390|M.avium_104 FAAAVMIGVALTGVVCFAGFFASYMFNVYFQNGAPGSFVSTSASFATTGD
MAB_1012c|M.abscessus_ATCC_199 FAAAIMIGIALTGVVCFVGFLGAYLFNVYWQNGAPGSFVTTFSAFATPGD
Mb0173|M.bovis_AF2122/97 VLASMLVATLLNGLVITVGLVGGFLFGVYLQNVSGGAYLATLTLITGLPE
Rv0167|M.tuberculosis_H37Rv VLASMLVATLLNGLVITVGLVGGFLFGVYLQNVSGGAYLATLTLITGLPE
MLBr_02587|M.leprae_Br4923 VLAATLVATLLNGLVITVGLVGGYLFGVYLQNVSGGAYLATLTTITGLPE
. *: ::. *.*:* .*:...::*.** *: : *::::* : :: :
MSMEG_1141|M.smegmatis_MC2_155 MIVALIKAVIFGAIVAVVSCQKGLSTVGGPTGVANSVNAAVVESILLLMV
Mvan_0957|M.vanbaalenii_PYR-1 MIVALVKAVIFGAIVAVVSCQKGLATQGGPTGVANSVNAAVVESILLLMV
Mflv_0012|M.gilvum_PYR-GCK MIVALVKAVIFGAIVAIVSCQKGLSTQGGPTGVANSVNAAVVESILVLMV
TH_2930|M.thermoresistible__bu MLVALIKAVVFGAIVAVVSCQKGLSTKGGPTGVANSVNAAVVESILVLMI
MMAR_3862|M.marinum_M MFLALFKAVIFGAIVAVVSAQKGLSTKGGPTGVANSVNAAVVESILVLMI
MUL_3794|M.ulcerans_Agy99 MFLALFKAVMFGAIVAVVSAQKGLSTKGGPTGVANSVNAAVVESILVLMI
MAV_2390|M.avium_104 MILALLKAIVFGAIVAIVSSQKGLSTKGGPTGVANSVNAAVVEAILLLMI
MAB_1012c|M.abscessus_ATCC_199 MILALIKAVIYGAIVAVVACQKGLSTKGGPIGVANSVNAAVVESILLLMT
Mb0173|M.bovis_AF2122/97 VVIATIKAATFGLIAGLVGCYRGLTVRGGSKGLGTAVNETVVLCVIALFA
Rv0167|M.tuberculosis_H37Rv VVIATIKAATFGLIAGLVGCYRGLTVRGGSKGLGTAVNETVVLCVIALFA
MLBr_02587|M.leprae_Br4923 VVIATVKAATFGLIAGLVGCYRGLIVRGGSNGLGAAVNETVVLCVVALYA
:.:* .** :* *..:*.. :** . **. *:. :** :** .:: *
MSMEG_1141|M.smegmatis_MC2_155 VNVAISQLYIMLFPRVGL
Mvan_0957|M.vanbaalenii_PYR-1 VNVAISQLYIMMFPRVGL
Mflv_0012|M.gilvum_PYR-GCK VNVAISQLYILMFPRVGL
TH_2930|M.thermoresistible__bu VNVGISQLYIMLFPRVGL
MMAR_3862|M.marinum_M VNVSISQLHVMLAPRTGL
MUL_3794|M.ulcerans_Agy99 VNVSISQLHVMLAPRTGL
MAV_2390|M.avium_104 VNVAISQLYIMLSPRTGL
MAB_1012c|M.abscessus_ATCC_199 VNLAISQLYIMMFPRTGL
Mb0173|M.bovis_AF2122/97 VNVILTTIGVRFG--TGR
Rv0167|M.tuberculosis_H37Rv VNVILTTIGVRFG--TGR
MLBr_02587|M.leprae_Br4923 VNVVLTTIGVRFG--TGH
**: :: : : : .*