For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PAAQQRELQRAGTVHRVTTAVGDPAWLITGYATVRQLFDDERVGRSHPEPDTAARSGDSAFFGGPIGSYE TEREDHARGRRLMQPHFTPKQMRTLAGRVHELADELITAMIDRGSPADFYTAVAVPLPMMVLCELLGVPY ADRDEFREWTVAASNTRDRSRSEWGMGQLFVYGMQLVGRKRQQPGDDVISRLCTIDGVADHEIASQSMAL LLGGHETTVVQLGLATLLLLANPEQWALLVSQPDLVPNAVEETLRASRTGGGEIPRYAREDLEIDGVHIA AGELLLLDVGAANHDPAVFGCPDQLDVARKHLAHVIFGYGSRYCVGAPLARMQLTEVLGQLVERLPGLHL RRDISELTMRGDLLTGGLREVPVGW*
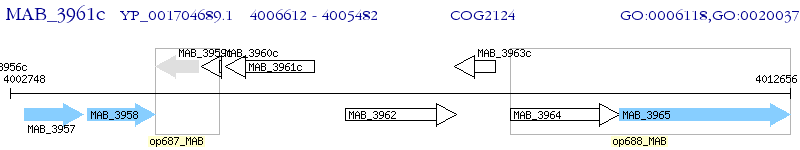
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3961c | - | - | 100% (376) | cytochrome P450 |
| M. abscessus ATCC 19977 | MAB_0276 | - | 2e-37 | 32.70% (315) | cytochrome P450 |
| M. abscessus ATCC 19977 | MAB_3825 | - | 5e-36 | 29.20% (387) | cytochrome P450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0801 | cyp126 | 4e-34 | 30.90% (356) | cytochrome P450 126 |
| M. gilvum PYR-GCK | Mflv_0225 | - | 6e-53 | 34.10% (393) | cytochrome P450 |
| M. tuberculosis H37Rv | Rv3518c | cyp142 | 2e-39 | 33.98% (309) | cytochrome P450 monooxygenase 142 |
| M. leprae Br4923 | MLBr_02088 | - | 4e-41 | 35.74% (319) | putative cytochrome p450 |
| M. marinum M | MMAR_4762 | cyp105Q4 | 5e-50 | 35.01% (377) | cytochrome P450 105Q4 Cyp105Q4 |
| M. avium 104 | MAV_2968 | - | 9e-52 | 34.72% (386) | cytochrome P450-SU2 |
| M. smegmatis MC2 155 | MSMEG_0762 | - | 3e-53 | 34.02% (388) | cytochrome P450 FAS1 |
| M. thermoresistible (build 8) | TH_4319 | - | 1e-54 | 35.81% (377) | PUTATIVE cytochrome P450 |
| M. ulcerans Agy99 | MUL_0333 | cyp105Q4 | 2e-49 | 34.75% (377) | cytochrome P450 105Q4 Cyp105Q4 |
| M. vanbaalenii PYR-1 | Mvan_0682 | - | 2e-50 | 33.33% (393) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4762|M.marinum_M ---------------MSDTLASPSPETASGIPDYPMSRSAGCPFAPPPGV
MUL_0333|M.ulcerans_Agy99 ---------------MSDTLASPSPETAAGIPDYPMSRSAGCPFAPPPGV
TH_4319|M.thermoresistible__bu -------------VNVSDTLTGTDADATAAIPEYPMERDGRCPFAPPPGV
MAV_2968|M.avium_104 -------------------MTQPSTDADTDIPDFPMTRAPGCPFAPPPKV
Mflv_0225|M.gilvum_PYR-GCK -------------------MSQATGEAAPDLPPLHMRRDG---FDPTPHL
Mvan_0682|M.vanbaalenii_PYR-1 -------------------MSQAV---RPELPPVHMRRDG---FDPTPQL
MSMEG_0762|M.smegmatis_MC2_155 -------------------MTQAQ-----ALPPLHIRRDA---FDPTPEL
MAB_3961c|M.abscessus_ATCC_199 --------------------------------------------MPAAQQ
MLBr_02088|M.leprae_Br4923 MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTRADP
Rv3518c|M.tuberculosis_H37Rv ------------------------------MTEAPDVDLADGNFYASREA
Mb0801|M.bovis_AF2122/97 -------------------MTTAAGLSGIDLTDLDNFADGFPHHLFAIHR
MMAR_4762|M.marinum_M MALAAAKPLTRVRIWDGSTPWLITGYEQVRELFSDSRVSVDDRLPGFPHW
MUL_0333|M.ulcerans_Agy99 MSLAAAKPLTRVRIWDGSAPWLITGYEQVRELFSGSRVSVDDRVPGFPHW
TH_4319|M.thermoresistible__bu MELAAAKPLSRVRIWDGSTPWLVTGYETARALFADSRVSVDDRRPGFPHW
MAV_2968|M.avium_104 LQLNADKQLSRVRIWDGSTPWLVHGYQAIRALFADARTSVDDRLPGYPHW
Mflv_0225|M.gilvum_PYR-GCK REIREREGVRSVTNAFGMQVYLFTRYDDIKTVLSDHSRFSNGRPPGFVVP
Mvan_0682|M.vanbaalenii_PYR-1 REIRETEGVRVITSAFGMSAYLVTRHEDVKTVLSDHTRFSNTRPPGFVVP
MSMEG_0762|M.smegmatis_MC2_155 GEIRAGEGVHVTVNPFGMQVYLVTRHEDVKTVLSDHERFSNSRPPGFVLP
MAB_3961c|M.abscessus_ATCC_199 RELQRAGTVHRVTTAVGDPAWLITGYATVRQLFDDERVGRSHPEP-----
MLBr_02088|M.leprae_Br4923 FPVYRALIDYGPMQLPGMPLTVFSSFSDCDEALRHPLSASDRLKATLAQQ
Rv3518c|M.tuberculosis_H37Rv RAAYRWMRANQPVFRDRNGLAAASTYQAVIDAERQPELFSN---AGGIRP
Mb0801|M.bovis_AF2122/97 REAPVYWHRPTEHTPDGEGFWSVATYAETLEVLRDPVTYSSVTGGQRRFG
. .
MMAR_4762|M.marinum_M NAGMLS---TVHKRPRSVFTADGEEHTRFRRMLSKPFTFKRVEALRPTIQ
MUL_0333|M.ulcerans_Agy99 NAGMLS---TVHKRPRSVFTADGEEHTRFRRMLSKPFTFKRVEALRPTIQ
TH_4319|M.thermoresistible__bu NEMMRS---TIYKRPRSVFTTDAEEHTKFRRMLSKPFTFKRVEGLRPAIQ
MAV_2968|M.avium_104 NEGMLA---TVHKRPRSVFTSDAEEHTRFRRMLSKPFTFKRVEALRPAVQ
Mflv_0225|M.gilvum_PYR-GCK GAPPVDEETQARARAGNLLGLDPPEHQRLRRMLTPEFTHRRMQRLRPRIA
Mvan_0682|M.vanbaalenii_PYR-1 GAPPIDEDEQARSRAGNLLGLDPPEHQRLRRMLTPEFTLRRMRRLQPRIA
MSMEG_0762|M.smegmatis_MC2_155 GAPQISAEEQASNRAGNLLGLDPPEHQRLRRMLTPEFTIRRIKRLEPRIV
MAB_3961c|M.abscessus_ATCC_199 DTAARSGDSAFFGGPIGSYETEREDHARGRRLMQPHFTPKQMRTLAGRVH
MLBr_02088|M.leprae_Br4923 AIAAGAEPRPFYAS--SFMFLDPPDHTRLRKLVSKAFAPKVVQALEGDIA
Rv3518c|M.tuberculosis_H37Rv DQPALP----------MMIDMDDPAHLLRRKLVNAGFTRKRVKDKEASIA
Mb0801|M.bovis_AF2122/97 GTVLQD----LPVAGQVLNMMDDPRHTRIRRLVSSGLTPRMIRRVEDDLR
: * *::: :: : :. :
MMAR_4762|M.marinum_M QITDEHIDAMLAGP---QPADLVAKLALPVPSLVISQLLGVPYEDAEMFQ
MUL_0333|M.ulcerans_Agy99 QITDEHIDAMLAGP---QPADLVAKLALPVPSLVISQLLGVPYEDAEMFQ
TH_4319|M.thermoresistible__bu KITDDHIDKILAGP---KPADLVTALALPVPSLVISEMLGVPYEDAEFFQ
MAV_2968|M.avium_104 KITDDHIDALLRGP---NPGDIVSTVSLPVPSLVISELLGVPYEDAEFFQ
Mflv_0225|M.gilvum_PYR-GCK EIVDTQIDALAAAG---RPADLVEHFALPIPSLVICELLGVPYADRDDFQ
Mvan_0682|M.vanbaalenii_PYR-1 EIVDAQLDALAAARDGEASADLVQHFALPIPSLVICELLGVPYADRDDFQ
MSMEG_0762|M.smegmatis_MC2_155 EIVDAHLDAMESAG---PPADIVADFALPIPSLVICELLGVPYEDRTDFQ
MAB_3961c|M.abscessus_ATCC_199 ELADELITAMIDRG---SPADFYTAVAVPLPMMVLCELLGVPYADRDEFR
MLBr_02088|M.leprae_Br4923 ALVDSLLDKGAAAG----QFDVIADLAFPLAVAVICRLLGVPYEDAPEFG
Rv3518c|M.tuberculosis_H37Rv ALCDTLIDAVCERG----ECDFVRDLAAPLPMAVIGDMLGVRPEQRDMFL
Mb0801|M.bovis_AF2122/97 RRARGLLDGVEPGA----PFDFVVEIAAELPMQMICILLGVPETDRHWLF
: *. .: :. :: :*** : :
MMAR_4762|M.marinum_M HHANVGLARYA-----------TGADTVKGAMSLHKYLAELVEAKMANPA
MUL_0333|M.ulcerans_Agy99 HHANVGLARYA-----------TGADTVKGAMSLHKYLAELVEAKMANPA
TH_4319|M.thermoresistible__bu EHANRGLDRYA-----------KAEDNMSKGMSLSQYLIKLVEAKMQNPA
MAV_2968|M.avium_104 TQAQRGMGRYA-----------TEEDTAQGAASLAKYLANLVRAKMQSPS
Mflv_0225|M.gilvum_PYR-GCK RRSARQLDLSI-----------PIPERLELQREGREYMAALVAGARRHPG
Mvan_0682|M.vanbaalenii_PYR-1 RRSARQLDLSI-----------PIPERIELAREGRAYMGSLVAGARTNPG
MSMEG_0762|M.smegmatis_MC2_155 QRSARQLDLSA-----------PMPERLELQRQGRAYMRGLVERSRTRPG
MAB_3961c|M.abscessus_ATCC_199 EWTVAASNTRD-----------RSRSEWGMG-QLFVYGMQLVGRKRQQPG
MLBr_02088|M.leprae_Br4923 RVSALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQLVKCRRGTPG
Rv3518c|M.tuberculosis_H37Rv RWSDDLVTFLSSHVS-----QEDFQITMDAFAAYNDFTRATIAARRADPT
Mb0801|M.bovis_AF2122/97 EAVEPGFDFRGSRR-----ATMPRLNVEDAGSRLYTYALELIAGKRAEPA
: : *
MMAR_4762|M.marinum_M EDAVSDLAERVKAGE----LSVKEAAQLGTGLLIAGHETTANMIGLGVLA
MUL_0333|M.ulcerans_Agy99 EDAVSDLAERVKAGE----LSVKEAAQLGTGLLIAGHETTANMIGLGVLA
TH_4319|M.thermoresistible__bu EDAVSDLAERVKAGE----IDVKEAAQLGTGLLIAGHETTANMIGLGVLA
MAV_2968|M.avium_104 EDLVSDLAERVNAEE----ISVREAAQLATGVLIAGHETTANMISLSVAA
Mflv_0225|M.gilvum_PYR-GCK DDILG-MLIREHGGE----LTDDELIGVASLLLLAGHETTSNMLALGVLA
Mvan_0682|M.vanbaalenii_PYR-1 DDILG-MLVREHGAE----LTDDELVGIAGLLLLAGHETTSNMLALGTLA
MSMEG_0762|M.smegmatis_MC2_155 DDILG-MLVREHGTE----LTDDELIGIAGLLLLAGHETTSNMLGLGVLA
MAB_3961c|M.abscessus_ATCC_199 DDVISRLCTIDG-------VADHEIASQSMALLLGGHETTVVQLGLATLL
MLBr_02088|M.leprae_Br4923 EDLISRLIELDESGDQ---LTEEEIIATCGLLLVAGHETTVNLIANAVLA
Rv3518c|M.tuberculosis_H37Rv DDLVSVLVSSEVDGER---LSDDELVMETLLILIGGDETTRHTLSGGTEQ
Mb0801|M.bovis_AF2122/97 DDMLSVVANATIDDPDAPALSDAELYLFFHLLFSAGAETTRNSIAGGLLA
:* :. : : * :: .* *** :. .
MMAR_4762|M.marinum_M LLVNPDQAGILRDAQDPKIVANAVEELLRYLSIIQNGQRRVAHEDIHIGG
MUL_0333|M.ulcerans_Agy99 LLVNPDQAGILRDAHDPKIVANAVEELLRYLSIIQNGQRRVAHEDIHIGG
TH_4319|M.thermoresistible__bu LLEHPEQADFLRSAEDPKAIATAIEELMRYLSIIQNGQRRVAVEDIEIAG
MAV_2968|M.avium_104 LLEHPDQRALLCDTDDPKVIATAVEELMRYLSIIQTGQRRIAIEDIEIGG
Mflv_0225|M.gilvum_PYR-GCK LLRHPDQLAAIR--DDPAAVAPAVEELLRWLSIVQTSIPRITTTDVEIAG
Mvan_0682|M.vanbaalenii_PYR-1 LLRHPEQLAAVR--EDPDAVAPAVEELLRWLSIVHTAIPRITTTDVEIAG
MSMEG_0762|M.smegmatis_MC2_155 LLRHPDQLACVR--DDPDAVGPAIEELLRWLSIVSTALPRITTTDVELAG
MAB_3961c|M.abscessus_ATCC_199 LLANPEQWALLVS--QPDLVPNAVEETLRASRTGGGEIPRYAREDLEIDG
MLBr_02088|M.leprae_Br4923 MLRNPSQWKALSS--NPQRAPLVVEETLRYDPAIH-LIGRVAAKDMTIGQ
Rv3518c|M.tuberculosis_H37Rv LLRNRDQWDLLQR--DPSLLPGAIEEMLRWTAPVK-NMCRVLTADTEFHG
Mb0801|M.bovis_AF2122/97 LAENPDQLQTLRS--DFELLPTAIEEIVRWTSPSP-SKRRTASRAVSLGG
: : .* : : .:** :* * :
MMAR_4762|M.marinum_M ETIRAGEGIIIDLAPANWDAHAFTEPDRLYLHRAGAERNVAFGYGRHQCV
MUL_0333|M.ulcerans_Agy99 ETIRAGEGIIIDLAPANWDAHAFTEPDRLYLHRAGAERNVAFGYGRHQCV
TH_4319|M.thermoresistible__bu ETIRAGEGIILDLAPANWDPATFPEPDRLDLQRSDAP-QLGFGYGRHQCV
MAV_2968|M.avium_104 ETIRAGEGIILDVAPANWDARQFPNPDRLDLRRQDGP-HVGFGYGRHQCV
Mflv_0225|M.gilvum_PYR-GCK VAIPAGQLVFASLPAGNRDPEFIEHPEVLDIGR-GAPGHLAFGHGVHHCL
Mvan_0682|M.vanbaalenii_PYR-1 VSIPAGQLVFASLPSGNRDDEFIERPEVFDITR-GAMGHLAFGHGVHHCL
MSMEG_0762|M.smegmatis_MC2_155 VTIPAGHLVFASLPAGNRDPEFIDDPDTLDIRR-GAPGHLAFGHGVHHCL
MAB_3961c|M.abscessus_ATCC_199 VHIAAGELLLLDVGAANHDPAVFGCPDQLDVAR-KHLAHVIFGYGSRYCV
MLBr_02088|M.leprae_Br4923 TTLTEGDTMVLLLAAANRDPAVYSRPDEFDPDR-PSSRHLAFAVGSHFCL
Rv3518c|M.tuberculosis_H37Rv TALCAGEKMMLLFESANFDEAVFCEPEKFDVQR-NPNSHLAFGFGTHFCL
Mb0801|M.bovis_AF2122/97 QPIEAGQKVVVWEGSANRDPSVFDRADEFDITR-KPNPHLGFGQGVHYCL
: *. :. ..* * .: : * :: *. * : *:
MMAR_4762|M.marinum_M GQQLARAELQIVYRTLLQRIPTLTLATALEDVPFKDDRLAYGVYELPVTW
MUL_0333|M.ulcerans_Agy99 GQQLARTELQIVYRTLLQRIPTLTLATALEDVPFKDDRLAYGVYELPVTW
TH_4319|M.thermoresistible__bu GMQLARAELQIVFPTLLRRIPTLKLATTIDEIPFKHDRLAYGVYELPVTW
MAV_2968|M.avium_104 GQQLARMELQIVLPTLLRRVPTLRLAAPLDELPFKHDALAYGLYELPVTW
Mflv_0225|M.gilvum_PYR-GCK GAPLARMEMAIAFPALFRRFPSLEPAEDFADVAFRSFHFIYGLKSLEVSW
Mvan_0682|M.vanbaalenii_PYR-1 GAPLARMEMQIAFPALLRRFPTLAPAGEFDDVPFRSFHFIYGLKSLEVTW
MSMEG_0762|M.smegmatis_MC2_155 GAPLARMEMRIALPALLRRFPTLALAEPFEDVRWRPFHFIYGLQSLAVAW
MAB_3961c|M.abscessus_ATCC_199 GAPLARMQLTEVLGQLVERLPGLHLRRDISELTMRGDLLTGGLREVPVGW
MLBr_02088|M.leprae_Br4923 GAALARLEATVTLSAISARFPQVQLAG---ELVYKPNVAMRGMSALPVQV
Rv3518c|M.tuberculosis_H37Rv GNQLARLELSLMTERVLRRLPDLRLVADDSVLPLRPANFVSGLESMPVVF
Mb0801|M.bovis_AF2122/97 GANLARLELRVLFEELLSRFGSVRVVEP---AEWTRSNRHTGIRHLVVEL
* *** : : *. : *: : *
MMAR_4762|M.marinum_M -------
MUL_0333|M.ulcerans_Agy99 -------
TH_4319|M.thermoresistible__bu -------
MAV_2968|M.avium_104 -------
Mflv_0225|M.gilvum_PYR-GCK -------
Mvan_0682|M.vanbaalenii_PYR-1 -------
MSMEG_0762|M.smegmatis_MC2_155 -------
MAB_3961c|M.abscessus_ATCC_199 -------
MLBr_02088|M.leprae_Br4923 -------
Rv3518c|M.tuberculosis_H37Rv TPSPPLG
Mb0801|M.bovis_AF2122/97 RGG----