For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSQATGEAAPDLPPLHMRRDGFDPTPHLREIREREGVRSVTNAFGMQVYLFTRYDDIKTVLSDHSRFSNG RPPGFVVPGAPPVDEETQARARAGNLLGLDPPEHQRLRRMLTPEFTHRRMQRLRPRIAEIVDTQIDALAA AGRPADLVEHFALPIPSLVICELLGVPYADRDDFQRRSARQLDLSIPIPERLELQREGREYMAALVAGAR RHPGDDILGMLIREHGGELTDDELIGVASLLLLAGHETTSNMLALGVLALLRHPDQLAAIRDDPAAVAPA VEELLRWLSIVQTSIPRITTTDVEIAGVAIPAGQLVFASLPAGNRDPEFIEHPEVLDIGRGAPGHLAFGH GVHHCLGAPLARMEMAIAFPALFRRFPSLEPAEDFADVAFRSFHFIYGLKSLEVSW
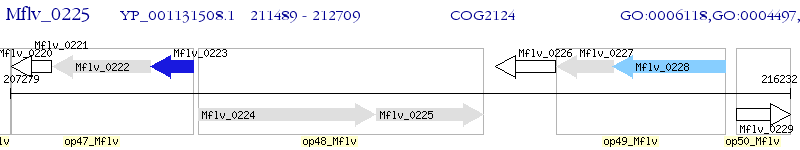
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0225 | - | - | 100% (406) | cytochrome P450 |
| M. gilvum PYR-GCK | Mflv_2418 | - | 8e-62 | 35.27% (414) | cytochrome P450 |
| M. gilvum PYR-GCK | Mflv_1229 | - | 1e-54 | 37.83% (341) | cytochrome P450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1912c | cyp140 | 1e-43 | 36.96% (303) | cytochrome p450 140 CYP140 |
| M. tuberculosis H37Rv | Rv3121 | cyp141 | 6e-47 | 32.06% (393) | cytochrome P450 141 |
| M. leprae Br4923 | MLBr_02088 | - | 4e-50 | 36.62% (355) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_3961c | - | 6e-53 | 34.10% (393) | cytochrome P450 |
| M. marinum M | MMAR_4762 | cyp105Q4 | 2e-64 | 37.65% (409) | cytochrome P450 105Q4 Cyp105Q4 |
| M. avium 104 | MAV_2464 | - | 2e-72 | 41.38% (406) | cytochrome P450 family protein |
| M. smegmatis MC2 155 | MSMEG_0762 | - | 1e-180 | 77.14% (398) | cytochrome P450 FAS1 |
| M. thermoresistible (build 8) | TH_4319 | - | 5e-70 | 37.75% (408) | PUTATIVE cytochrome P450 |
| M. ulcerans Agy99 | MUL_0333 | cyp105Q4 | 5e-64 | 37.41% (409) | cytochrome P450 105Q4 Cyp105Q4 |
| M. vanbaalenii PYR-1 | Mvan_0682 | - | 0.0 | 82.64% (409) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0225|M.gilvum_PYR-GCK -------------------MSQATGEAAPDLPPLHMRRDG---FDPTPHL
Mvan_0682|M.vanbaalenii_PYR-1 -------------------MSQAV---RPELPPVHMRRDG---FDPTPQL
MSMEG_0762|M.smegmatis_MC2_155 -------------------MTQAQ-----ALPPLHIRRDA---FDPTPEL
MMAR_4762|M.marinum_M ---------------MSDTLASPSPETASGIPDYPMSRSAGCPFAPPPGV
MUL_0333|M.ulcerans_Agy99 ---------------MSDTLASPSPETAAGIPDYPMSRSAGCPFAPPPGV
TH_4319|M.thermoresistible__bu -------------VNVSDTLTGTDADATAAIPEYPMERDGRCPFAPPPGV
MAV_2464|M.avium_104 ---------------MSISFETSESRADAELPVLPMPRAAHCPLAPPPEF
MAB_3961c|M.abscessus_ATCC_199 --------------------------------------------MPAAQQ
Mb1912c|M.bovis_AF2122/97 ---------MKDKLHWLAMHGVIRGIAAIGIRRGDLQARLIADPAVATDP
MLBr_02088|M.leprae_Br4923 MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTRADP
Rv3121|M.tuberculosis_H37Rv ---------------------------MTSTSIPTFPFDRPVPTEPSPML
.
Mflv_0225|M.gilvum_PYR-GCK REIREREGVRSVTNAFGMQVYLFTRYDDIKTVLSDHSRFSNGRPPGFVVP
Mvan_0682|M.vanbaalenii_PYR-1 REIRETEGVRVITSAFGMSAYLVTRHEDVKTVLSDHTRFSNTRPPGFVVP
MSMEG_0762|M.smegmatis_MC2_155 GEIRAGEGVHVTVNPFGMQVYLVTRHEDVKTVLSDHERFSNSRPPGFVLP
MMAR_4762|M.marinum_M MALAAAKPLTRVRIWDGSTPWLITGYEQVRELFSDSRVSVDDRLPGFPHW
MUL_0333|M.ulcerans_Agy99 MSLAAAKPLTRVRIWDGSAPWLITGYEQVRELFSGSRVSVDDRVPGFPHW
TH_4319|M.thermoresistible__bu MELAAAKPLSRVRIWDGSTPWLVTGYETARALFADSRVSVDDRRPGFPHW
MAV_2464|M.avium_104 VDWRQQPGLRRA-LFQGNPVWVVSRYHDIRAALVDPRLSAKT-IP-----
MAB_3961c|M.abscessus_ATCC_199 RELQRAGTVHRVTTAVGDPAWLITGYATVRQLFDDERVGRSHPEP-----
Mb1912c|M.bovis_AF2122/97 VPFYDEVRSHGALVRN------RANYLTVDHRLAHDLLRSDDFRVVSFGE
MLBr_02088|M.leprae_Br4923 FPVYRALIDYGPMQLPGMPLTVFSSFSDCDEALRHPLSASDRLKATLAQQ
Rv3121|M.tuberculosis_H37Rv SELRNSCPVAPIELPSGHTAWLVTRFDDVKGVLSDKRFSCRAAAHPSSPP
: . :
Mflv_0225|M.gilvum_PYR-GCK GAPPVDE-------ETQARARAGNLLG-LDPPEHQRLRRMLTPEFTHRRM
Mvan_0682|M.vanbaalenii_PYR-1 GAPPIDE-------DEQARSRAGNLLG-LDPPEHQRLRRMLTPEFTLRRM
MSMEG_0762|M.smegmatis_MC2_155 GAPQISA-------EEQASNRAGNLLG-LDPPEHQRLRRMLTPEFTIRRI
MMAR_4762|M.marinum_M NAGMLS----------TVHKRPRSVFT-ADGEEHTRFRRMLSKPFTFKRV
MUL_0333|M.ulcerans_Agy99 NAGMLS----------TVHKRPRSVFT-ADGEEHTRFRRMLSKPFTFKRV
TH_4319|M.thermoresistible__bu NEMMRS----------TIYKRPRSVFT-TDAEEHTKFRRMLSKPFTFKRV
MAV_2464|M.avium_104 DSIMPT----------DADNKVPVMFARTDDPEHHRLRRMLTGNFTFRRC
MAB_3961c|M.abscessus_ATCC_199 DTAARSG-------DSAFFGGPIGSYE-TEREDHARGRRLMQPHFTPKQM
Mb1912c|M.bovis_AF2122/97 NLPPPLRWLERRTRGDQLHPLREPSLLAVEPPDHTRYRKTVSAVFTSRAV
MLBr_02088|M.leprae_Br4923 AIAA----------GAEPRPFYASSFMFLDPPDHTRLRKLVSKAFAPKVV
Rv3121|M.tuberculosis_H37Rv ----FVP-----------FVQLCPSLLSIDGPQHTAARRLLAQGLNPGFI
: :* *: : :
Mflv_0225|M.gilvum_PYR-GCK QRLRPRIAEIVDTQIDALAAAG---RPADLVEHFALPIPSLVICELLGVP
Mvan_0682|M.vanbaalenii_PYR-1 RRLQPRIAEIVDAQLDALAAARDGEASADLVQHFALPIPSLVICELLGVP
MSMEG_0762|M.smegmatis_MC2_155 KRLEPRIVEIVDAHLDAMESAG---PPADIVADFALPIPSLVICELLGVP
MMAR_4762|M.marinum_M EALRPTIQQITDEHIDAMLAGP---QPADLVAKLALPVPSLVISQLLGVP
MUL_0333|M.ulcerans_Agy99 EALRPTIQQITDEHIDAMLAGP---QPADLVAKLALPVPSLVISQLLGVP
TH_4319|M.thermoresistible__bu EGLRPAIQKITDDHIDKILAGP---KPADLVTALALPVPSLVISEMLGVP
MAV_2464|M.avium_104 ESMRPQIQDTVDHYLDRMVDGG---VPADLVREFALPVPSLVIALLLGVP
MAB_3961c|M.abscessus_ATCC_199 RTLAGRVHELADELITAMIDRG---SPADFYTAVAVPLPMMVLCELLGVP
Mb1912c|M.bovis_AF2122/97 SALRDLVEQTAINLLDRFAEQP---GIVDVVGRYCSQLPIVVISEILGVP
MLBr_02088|M.leprae_Br4923 QALEGDIAALVDSLLDKGAA-A---GQFDVIADLAFPLAVAVICRLLGVP
Rv3121|M.tuberculosis_H37Rv ARMRPVVQQIVDNALDDLAAAEP---PVDFQEIVSVPIGEQLMAKLLGVE
: : . : *. . : ::. :***
Mflv_0225|M.gilvum_PYR-GCK YADRDDFQRRSARQLDLSI-----------PIPERLELQREGREYMAALV
Mvan_0682|M.vanbaalenii_PYR-1 YADRDDFQRRSARQLDLSI-----------PIPERIELAREGRAYMGSLV
MSMEG_0762|M.smegmatis_MC2_155 YEDRTDFQQRSARQLDLSA-----------PMPERLELQRQGRAYMRGLV
MMAR_4762|M.marinum_M YEDAEMFQHHANVGLARYA-----------TGADTVKGAMSLHKYLAELV
MUL_0333|M.ulcerans_Agy99 YEDAEMFQHHANVGLARYA-----------TGADTVKGAMSLHKYLAELV
TH_4319|M.thermoresistible__bu YEDAEFFQEHANRGLDRYA-----------KAEDNMSKGMSLSQYLIKLV
MAV_2464|M.avium_104 PEDLELFQFNTTKGLDQKS-----------SDEEKGKAFGAMYAYIEELV
MAB_3961c|M.abscessus_ATCC_199 YADRDEFREWTVAASNTRD-----------RSRSEWGMG-QLFVYGMQLV
Mb1912c|M.bovis_AF2122/97 EHDRPRVLEFGELAAPSLD--IGIPWRQYLRVQQGIRG---FDCWLEGHL
MLBr_02088|M.leprae_Br4923 YEDAPEFGRVSALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQLV
Rv3121|M.tuberculosis_H37Rv PKTVHELAAHVDAAMSVCEIG----------DEEVSRRWSALCTMVIDIL
. . :
Mflv_0225|M.gilvum_PYR-GCK AGARRHPGDDILG--MLIREHGG---ELTDDELIGVASLLLLAGHETTSN
Mvan_0682|M.vanbaalenii_PYR-1 AGARTNPGDDILG--MLVREHGA---ELTDDELVGIAGLLLLAGHETTSN
MSMEG_0762|M.smegmatis_MC2_155 ERSRTRPGDDILG--MLVREHGT---ELTDDELIGIAGLLLLAGHETTSN
MMAR_4762|M.marinum_M EAKMANPAEDAVS-DLAERVKAG---ELSVKEAAQLGTGLLIAGHETTAN
MUL_0333|M.ulcerans_Agy99 EAKMANPAEDAVS-DLAERVKAG---ELSVKEAAQLGTGLLIAGHETTAN
TH_4319|M.thermoresistible__bu EAKMQNPAEDAVS-DLAERVKAG---EIDVKEAAQLGTGLLIAGHETTAN
MAV_2464|M.avium_104 QRKAREPGDDLISRLITEYVATG---QLDHATTAMNSVIMMQAGHETTAN
MAB_3961c|M.abscessus_ATCC_199 GRKRQQPGDDVIS----RLCTID---GVADHEIASQSMALLLGGHETTVV
Mb1912c|M.bovis_AF2122/97 QQLRHAPGDDLMSQLIQIAESGDNETQLDETELRAIAGLVLVAGFETTVN
MLBr_02088|M.leprae_Br4923 KCRRGTPGEDLISRLIELDESGD---QLTEEEIIATCGLLLVAGHETTVN
Rv3121|M.tuberculosis_H37Rv HRKLAEPGDDLLSTIAQANRQQS---TMTDEQVVGMLLTVVIGGVDTPIA
*.:* :. : :: .* :*.
Mflv_0225|M.gilvum_PYR-GCK MLALGVLALLRHPDQLAAIR--DDPAAVAPAVEELLRWLSIVQTSIPRIT
Mvan_0682|M.vanbaalenii_PYR-1 MLALGTLALLRHPEQLAAVR--EDPDAVAPAVEELLRWLSIVHTAIPRIT
MSMEG_0762|M.smegmatis_MC2_155 MLGLGVLALLRHPDQLACVR--DDPDAVGPAIEELLRWLSIVSTALPRIT
MMAR_4762|M.marinum_M MIGLGVLALLVNPDQAGILRDAQDPKIVANAVEELLRYLSIIQNGQRRVA
MUL_0333|M.ulcerans_Agy99 MIGLGVLALLVNPDQAGILRDAHDPKIVANAVEELLRYLSIIQNGQRRVA
TH_4319|M.thermoresistible__bu MIGLGVLALLEHPEQADFLRSAEDPKAIATAIEELMRYLSIIQNGQRRVA
MAV_2464|M.avium_104 MISLGTVALLGHPEIYARLGQTDDSAVVANIVEELMRYLSIVHSQVDRVA
MAB_3961c|M.abscessus_ATCC_199 QLGLATLLLLANPEQWALLVS--QPDLVPNAVEETLRASRTGGGEIPRYA
Mb1912c|M.bovis_AF2122/97 LLGNGIRMLLDTPEHLATLRQ--HPELWPNTVEEILRLDSPVQ-LTARVA
MLBr_02088|M.leprae_Br4923 LIANAVLAMLRNPSQWKALSS--NPQRAPLVVEETLRYDPAIH-LIGRVA
Rv3121|M.tuberculosis_H37Rv VITNGLASLLHHRDQYERLVE--DPGRVARAVEEIVRFNPATEIEHLRVV
: . :* . : .. :** :* * .
Mflv_0225|M.gilvum_PYR-GCK TTDVEIAGVAIPAGQLVFASLPAGNRDPEFIEHPEVLDIGR-GAPGHLAF
Mvan_0682|M.vanbaalenii_PYR-1 TTDVEIAGVSIPAGQLVFASLPSGNRDDEFIERPEVFDITR-GAMGHLAF
MSMEG_0762|M.smegmatis_MC2_155 TTDVELAGVTIPAGHLVFASLPAGNRDPEFIDDPDTLDIRR-GAPGHLAF
MMAR_4762|M.marinum_M HEDIHIGGETIRAGEGIIIDLAPANWDAHAFTEPDRLYLHRAGAERNVAF
MUL_0333|M.ulcerans_Agy99 HEDIHIGGETIRAGEGIIIDLAPANWDAHAFTEPDRLYLHRAGAERNVAF
TH_4319|M.thermoresistible__bu VEDIEIAGETIRAGEGIILDLAPANWDPATFPEPDRLDLQRSDAP-QLGF
MAV_2464|M.avium_104 TEGLTIAGQLIRAGEFVVMNLPAGNWDTEFVDNPESFDADR-NPRGHLGF
MAB_3961c|M.abscessus_ATCC_199 REDLEIDGVHIAAGELLLLDVGAANHDPAVFGCPDQLDVAR-KHLAHVIF
Mb1912c|M.bovis_AF2122/97 CRDVEVAGVRIKRGEVVVIYLAAANRDPAVFPDPHRFDIERPNAGRHLAF
MLBr_02088|M.leprae_Br4923 AKDMTIGQTTLTEGDTMVLLLAAANRDPAVYSRPDEFDPDRPSS-RHLAF
Rv3121|M.tuberculosis_H37Rv TEDVVIAGTALSAGSPAFTSITSANRDSDQFLDPDEFDVER-NPNEHIAF
.: : : * . : ..* * *. : * :: *
Mflv_0225|M.gilvum_PYR-GCK GHGVHHCLGAPLARMEMAIAFPALFRRFPSLEPAEDFADVAFRSFHFIYG
Mvan_0682|M.vanbaalenii_PYR-1 GHGVHHCLGAPLARMEMQIAFPALLRRFPTLAPAGEFDDVPFRSFHFIYG
MSMEG_0762|M.smegmatis_MC2_155 GHGVHHCLGAPLARMEMRIALPALLRRFPTLALAEPFEDVRWRPFHFIYG
MMAR_4762|M.marinum_M GYGRHQCVGQQLARAELQIVYRTLLQRIPTLTLATALEDVPFKDDRLAYG
MUL_0333|M.ulcerans_Agy99 GYGRHQCVGQQLARTELQIVYRTLLQRIPTLTLATALEDVPFKDDRLAYG
TH_4319|M.thermoresistible__bu GYGRHQCVGMQLARAELQIVFPTLLRRIPTLKLATTIDEIPFKHDRLAYG
MAV_2464|M.avium_104 GYGVHQCIGANLARVEMQVAFATLARRLPGLRLAVPPEQLKFK-DANIYG
MAB_3961c|M.abscessus_ATCC_199 GYGSRYCVGAPLARMQLTEVLGQLVERLPGLHLRRDISELTMRGDLLTGG
Mb1912c|M.bovis_AF2122/97 STGRHFCLGAALARAEGEVGLRTFFDRFPDVRAAG---AGSRRDTRVLRG
MLBr_02088|M.leprae_Br4923 AVGSHFCLGAALARLEATVTLSAISARFPQVQLAG---ELVYKPNVAMRG
Rv3121|M.tuberculosis_H37Rv GYGPHACPASAYSRMCLTTFFTSLTQRFPQLQLARPFEDLERRGKGLHSV
. * : * . :* : *:* : :
Mflv_0225|M.gilvum_PYR-GCK -LKSLEVSW---------
Mvan_0682|M.vanbaalenii_PYR-1 -LKSLEVTW---------
MSMEG_0762|M.smegmatis_MC2_155 -LQSLAVAW---------
MMAR_4762|M.marinum_M -VYELPVTW---------
MUL_0333|M.ulcerans_Agy99 -VYELPVTW---------
TH_4319|M.thermoresistible__bu -VYELPVTW---------
MAV_2464|M.avium_104 -MKELPVSW---------
MAB_3961c|M.abscessus_ATCC_199 -LREVPVGW---------
Mb1912c|M.bovis_AF2122/97 -WSTLPVTLGPARSMVSP
MLBr_02088|M.leprae_Br4923 -MSALPVQV---------
Rv3121|M.tuberculosis_H37Rv GIKELLVTWPT-------
: *