For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPITTDPGTLLLQVLDPANRANPYPVYGQLVEQTQAGPVHPPGMDVSVLATFADCNAVLRHPQASSDRRK SNIVQRQLAQNPNLIARPSFLGLDAPDHTRLRKLASKAFAPRVINAMAEDIQTFVDKMLDDIALRGTFNL VTEFAYPLPVAVICRMLGVPIEDEPEFGRASALLGQGLDPVYALTGQSPANMDERFQAARWMWDYFIDLI ASRRRNLGDDLLSALIQVEEAGDQLTEEEIISTCTLLLIAGHETTVNLIANASLAMLRHPEQWKLLGQNA DRAPLVVEETLRYDPPVHMVARVADGEMAIRDFILPDGEWVLLMLAAAQRDPDVGPDLDVFNPDRSEIKH LAFGHGPHFCLGAPLARLEARLALSSITARFPGARLMDEPTYKPNVTLRGLAELPVSIA
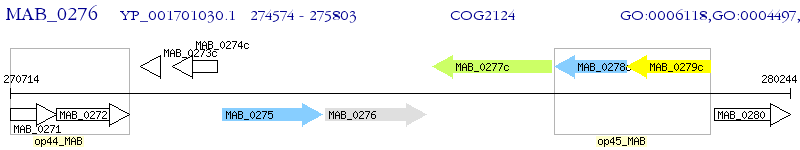
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0276 | - | - | 100% (409) | cytochrome P450 |
| M. abscessus ATCC 19977 | MAB_4456 | - | 6e-53 | 38.44% (333) | putative cytochrome P450 |
| M. abscessus ATCC 19977 | MAB_0917c | - | 2e-51 | 33.00% (397) | cytochrome P450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1912c | cyp140 | 3e-47 | 36.97% (330) | cytochrome p450 140 CYP140 |
| M. gilvum PYR-GCK | Mflv_1229 | - | 1e-128 | 55.37% (410) | cytochrome P450 |
| M. tuberculosis H37Rv | Rv1880c | cyp140 | 4e-47 | 36.97% (330) | cytochrome p450 140 CYP140 |
| M. leprae Br4923 | MLBr_02088 | - | 1e-123 | 53.79% (409) | putative cytochrome p450 |
| M. marinum M | MMAR_5268 | cyp164A3 | 1e-126 | 54.93% (406) | cytochrome P450 164A3 Cyp164A3 |
| M. avium 104 | MAV_0358 | - | 1e-125 | 57.61% (401) | cytochrome P450 |
| M. smegmatis MC2 155 | MSMEG_6312 | - | 1e-124 | 57.21% (402) | cytochrome P450 107B1 |
| M. thermoresistible (build 8) | TH_1650 | - | 3e-70 | 53.15% (254) | cytochrome P450 107B1 |
| M. ulcerans Agy99 | MUL_0475 | cyp123A3 | 4e-50 | 33.42% (404) | cytochrome P450 123A3 Cyp123A3 |
| M. vanbaalenii PYR-1 | Mvan_5579 | - | 1e-124 | 54.27% (422) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1912c|M.bovis_AF2122/97 ----------------MKDKLHWLAMHGVIRGIAAIGIRRGDLQARLIAD
Rv1880c|M.tuberculosis_H37Rv ----------------MKDKLHWLAMHGVIRGIAAIGIRRGDLQARLIAD
Mflv_1229|M.gilvum_PYR-GCK ----------------------------------MTTAAEPQSLLLQMLD
Mvan_5579|M.vanbaalenii_PYR-1 ----------------------------------MTTAAEPQSLLLQMLA
MSMEG_6312|M.smegmatis_MC2_155 ------------------------MHNGWMS--TAATAQEAQGLLLQLLD
MMAR_5268|M.marinum_M MLRGPVTAATHFDHTFFMRRHNVSMTTRPIEPIEFTEQVEPQAFLFQLLD
MAV_0358|M.avium_104 ------------------------MTTAP------TESAESQGLLLQLLD
MLBr_02088|M.leprae_Br4923 -------MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLID
MAB_0276|M.abscessus_ATCC_1997 ----------------------------------MPITTDPGTLLLQVLD
TH_1650|M.thermoresistible__bu ----------------------------------MTVEADPQILLMQMLD
MUL_0475|M.ulcerans_Agy99 ----------------------------------MSVHVGGPELVLDPYD
Mb1912c|M.bovis_AF2122/97 PAVATDPVPFYDEVRSHG--ALVRNRANYLTVDHRLAHDLLRSDDFRVVS
Rv1880c|M.tuberculosis_H37Rv PAVATDPVPFYDEVRSHG--ALVRNRANYLTVDHRLAHDLLRSDDFRVVS
Mflv_1229|M.gilvum_PYR-GCK PANRADPYPLYARIRAQ---GPVQLPANNLTVFSTYAA---CDEILRHPH
Mvan_5579|M.vanbaalenii_PYR-1 PEVRADPYPVYAQILAH---GPMQLPGNNLTVFSSYAD---CDEVLRHPD
MSMEG_6312|M.smegmatis_MC2_155 PATRADPYPIYDRIRRG---GPLALPEANLAVFSSFSD---CDDVLRHPS
MMAR_5268|M.marinum_M PAIRADPYPLCARLRDH---GPLHLPEANFVLFSSFGD---CDEVLRHPS
MAV_0358|M.avium_104 PANRADPYRLYAQFRER---GALQLPEANLAVFLSYRD---CDEVLRHPS
MLBr_02088|M.leprae_Br4923 PGTRADPFPVYRALIDY---GPMQLPGMPLTVFSSFSD---CDEALRHPL
MAB_0276|M.abscessus_ATCC_1997 PANRANPYPVYGQLVEQTQAGPVHPPGMDVSVLATFAD---CNAVLRHPQ
TH_1650|M.thermoresistible__bu PANRSDPYPVYRRIRER---GPVQPAGGNVTVFSSYAD---CDAVLRHPD
MUL_0475|M.ulcerans_Agy99 YDFHEDPYPYYRRLRNEAP--LYHNQELGFWALSRRRD---VHQGFRNST
:* . . :*
Mb1912c|M.bovis_AF2122/97 FGENLPPPLRWLERRTRGDQLHPLREPSLLAVEPPDHTRYRKTVSAVFTS
Rv1880c|M.tuberculosis_H37Rv FGENLPPPLRWLERRTRGDQLHPLREPSLLAVEPPDHTRYRKTVSAVFTS
Mflv_1229|M.gilvum_PYR-GCK ASSDRLKSTLAQRAIADGAQPRPFGPPGFLFLDPPDHTRLRRLVSKAFVP
Mvan_5579|M.vanbaalenii_PYR-1 ASSDRLKSTAAQRAIADGAEARPFGPPGFLFLDPPDHTRLRWLVSKAFVP
MSMEG_6312|M.smegmatis_MC2_155 SCSDRTKSTIFQRQLAAETQPRPQGPASFLFLDPPDHTRLRGLVSKAFAP
MMAR_5268|M.marinum_M SCSDQTKSTVAQRRVEEEPALGRDFPPGFLFLDPPDHTRLRKLVSKAFAP
MAV_0358|M.avium_104 SSSDNVNSTVAKRQAAAGTAPVRQGPPGFLFLDPPDHTRLRRLVSKAFAP
MLBr_02088|M.leprae_Br4923 SASDRLKATLAQQAIAAGAEPRPFYASSFMFLDPPDHTRLRKLVSKAFAP
MAB_0276|M.abscessus_ATCC_1997 ASSDRRKSNIVQRQLAQN--PNLIARPSFLGLDAPDHTRLRKLASKAFAP
TH_1650|M.thermoresistible__bu SCSDGLKSTITRRQLAEGKDVRPLGPPGFLFLDPPDHTRLRGLVSKAFAP
MUL_0475|M.ulcerans_Agy99 TLSNRDGVSLDP----VSRGPHATKTMSFLAIDDPAHLRLRTLVSKGFTP
.: .:: :: * * * * .* *..
Mb1912c|M.bovis_AF2122/97 RAVSALRDLVEQTAINLLDRFAEQPGI---VDVVGRYCSQLPIVVISEIL
Rv1880c|M.tuberculosis_H37Rv RAVSALRDLVEQTAINLLDRFAEQPGI---VDVVGRYCSQLPIVVISEIL
Mflv_1229|M.gilvum_PYR-GCK KVVKALEPEIVALVDALL--ADAGGT----FDAIEGLAYPLPVAVICRLL
Mvan_5579|M.vanbaalenii_PYR-1 KVVKALEPDIVTMVDGLLREAEAGAT----FEAIGGLAYPLPVAVICRLL
MSMEG_6312|M.smegmatis_MC2_155 RVIKRLEPEITALVDQLLD-AVDGPE----FNLIDNLAYPLPVAVICRLL
MMAR_5268|M.marinum_M KVVNALRPEISSLVDGLLDRIAEQGR----FDVVSDFAYPLPVAVICRLL
MAV_0358|M.avium_104 RVVAALEPDIRSLVDGLLDRAADRGE----LEIVEDFAYPLPVAVICRLL
MLBr_02088|M.leprae_Br4923 KVVQALEGDIAALVDSLLDKGAAAGQ----FDVIADLAFPLAVAVICRLL
MAB_0276|M.abscessus_ATCC_1997 RVINAMAEDIQTFVDKMLDDIALRGT----FNLVTEFAYPLPVAVICRML
TH_1650|M.thermoresistible__bu KVVRALEADIAAMVDELLAAAEPGGEP---FDVISGLAYPLPVTVICRLL
MUL_0475|M.ulcerans_Agy99 RRIRELEPRVTELAVAHLDTMLDMAGGSQTVDYVAEFAGKLPMDVISELM
: : : : . * .: : . *.: **..::
Mb1912c|M.bovis_AF2122/97 GVPEHDRPRVLEFGELAAPSLD-----IGIPWRQYLRVQQGIRGFDCWLE
Rv1880c|M.tuberculosis_H37Rv GVPEHDRPRVLEFGELAAPSLD-----IGIPWRQYLRVQQGIRGFDCWLE
Mflv_1229|M.gilvum_PYR-GCK GVPLADEPEFSAASALLAQSLDPFATVTGSAGDGFEERMEAARWLRGYLR
Mvan_5579|M.vanbaalenii_PYR-1 GVPLEDEPQFSAASALLAQSLDPFISVTGSAGSGFEERMQAARWLRGYLR
MSMEG_6312|M.smegmatis_MC2_155 GVPIEDEPKFSRASALLAAALDPFLALTGETSDLFDEQMKAGMWLRDYLR
MMAR_5268|M.marinum_M GVPLEDEPQFSQASALLAQALDPFFAFTGQNAENLNDSLEAGQWLRGYLR
MAV_0358|M.avium_104 GVPLDDEPQFSRASALLAQALDPFSTITGVPAEVASERQQAGTWLRDYFH
MLBr_02088|M.leprae_Br4923 GVPYEDAPEFGRVSALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLE
MAB_0276|M.abscessus_ATCC_1997 GVPIEDEPEFGRASALLGQGLDPVYALTGQSPANMDERFQAARWMWDYFI
TH_1650|M.thermoresistible__bu GVPIEDEPRFRDASALLAQGLDPFLTVTGQAADGHEQRVAAGQWLRGYLG
MUL_0475|M.ulcerans_Agy99 GVPVADRDQIRAMADGVMHRED-------GVTDVPASAIEASINLIVYYQ
*** * .. . * . : :
Mb1912c|M.bovis_AF2122/97 GHLQQLRHAPGDDLMSQLIQIAESGDNETQLDETELRAIAGLVLVAGFET
Rv1880c|M.tuberculosis_H37Rv GHLQQLRHAPGDDLMSQLIQIAESGDNETQLDETELRAIAGLVLVAGFET
Mflv_1229|M.gilvum_PYR-GCK ELISQRRRDPGEDLMSGLIQVEESGD---QLTEDEIVATCNLLLVAGHET
Mvan_5579|M.vanbaalenii_PYR-1 ELIARRRRDPGEDLMSGLIHVEESGD---QLTEDEIVATCNLLLVAGHET
MSMEG_6312|M.smegmatis_MC2_155 ALIDERRRTPGEDLMSGLVAVEESGD---QLTEDEIIATCNLLLIAGHET
MMAR_5268|M.marinum_M DLIALRRSQPGEDLMSGLVAVEESGD---QLTEDEIVSTCVLLLVAGHET
MAV_0358|M.avium_104 QLIEARRSRPGDDLLSGLIAVEESGD---QLTEEEIVSTCNLLLIAGHET
MLBr_02088|M.leprae_Br4923 QLVKCRRGTPGEDLISRLIELDESGD---QLTEEEIIATCGLLLVAGHET
MAB_0276|M.abscessus_ATCC_1997 DLIASRRRNLGDDLLSALIQVEEAGD---QLTEEEIISTCTLLLIAGHET
TH_1650|M.thermoresistible__bu ELLEARRADPRDDLMSALIEVVESGD---QLSTEEIIATCNLLLVAGXXX
MUL_0475|M.ulcerans_Agy99 QMIAERRKKPSDDLTSALLAADVDGD---RLTDEEVLGFMFLMVIAGNET
: * :** * *: ** :* *: . *:::**
Mb1912c|M.bovis_AF2122/97 TVNLLGNGIRMLLDTPEHLATLRQHPELWPNTVEEILRLDSPVQLTARVA
Rv1880c|M.tuberculosis_H37Rv TVNLLGNGIRMLLDTPEHLATLRQHPELWPNTVEEILRLDSPVQLTARVA
Mflv_1229|M.gilvum_PYR-GCK TVNLIANAILALLSNDGQWSALAADPARVGAVVEETLRYDPPVQLVGRIA
Mvan_5579|M.vanbaalenii_PYR-1 TVNLIANAILALLRNDGQWAALAADPGRAAAVIEETLRHDPPVQLVGRIA
MSMEG_6312|M.smegmatis_MC2_155 TVNLIANAALAMLRTPGQWAALAADGSRASAVIEETMRYDPPVQLVSRYA
MMAR_5268|M.marinum_M TVNLIGNAILAMLRDRRQWAQLGADPDRAAAVVEETMRYDPPVQLIGRIA
MAV_0358|M.avium_104 TVNLIGNAVLAMLRDPGQWAALGADPGRAPAIVEETLRYDPPVQLAGQIA
MLBr_02088|M.leprae_Br4923 TVNLIANAVLAMLRNPSQWKALSSNPQRAPLVVEETLRYDPAIHLIGRVA
MAB_0276|M.abscessus_ATCC_1997 TVNLIANASLAMLRHPEQWKLLGQNADRAPLVVEETLRYDPPVHMVARVA
TH_1650|M.thermoresistible__bu -----------------AGGGAGPGPRRG-------QRTGGADHCADRCA
MUL_0475|M.ulcerans_Agy99 TTKLLANAAFWGHKNTDQLTPIYADLSRVPLWVEETLRYDTSSQILARTV
* . . : : .
Mb1912c|M.bovis_AF2122/97 CRDVEVAG-----------VRIKRGEVVVIYLAAANRDPAVFPDPHRFDI
Rv1880c|M.tuberculosis_H37Rv CRDVEVAG-----------VRIKRGEVVVIYLAAANRDPAVFPDPHRFDI
Mflv_1229|M.gilvum_PYR-GCK AEDMTVG-----------TTTVPKGDNMLLLTAAAHRDPDAVDRPDEFDP
Mvan_5579|M.vanbaalenii_PYR-1 GQDMTIGGERSDGGNVAGGVTVPKGDNILLLTAAAHRDPDHVDRPDEFDP
MSMEG_6312|M.smegmatis_MC2_155 GDDLTIG-----------THTVPKGDTMLLLLAAAHRDPTIVGAPDRFDP
MMAR_5268|M.marinum_M GADMVIG-----------DVEVAKGDVMMMLVAAAHRDPAEYDRPEEFDP
MAV_0358|M.avium_104 LDDMVIG-----------GVEVPAGDVMMLLLAAANRDPAEFDRPDTFDP
MLBr_02088|M.leprae_Br4923 AKDMTIG-----------QTTLTEGDTMVLLLAAANRDPAVYSRPDEFDP
MAB_0276|M.abscessus_ATCC_1997 DGEMAIR-----------DFILPDGEWVLLMLAAAQRDPDVGPDLDVFNP
TH_1650|M.thermoresistible__bu AP--------------------RRGRQ------GAGRDAHRTAR----NP
MUL_0475|M.ulcerans_Agy99 VGELTLYD-----------TTIPEGDVVLLLPGSAHRDERAFRDPDEYLI
* .* **
Mb1912c|M.bovis_AF2122/97 ERPNAGRHLAFSTGRHFCLGAALARAEGEVGLRTFFDRFPDVRAA-GAGS
Rv1880c|M.tuberculosis_H37Rv ERPNAGRHLAFSTGRHFCLGAALARAEGEVGLRTFFDRFPDVRAA-GAGS
Mflv_1229|M.gilvum_PYR-GCK DRESF-RHLGFGKGPHFCIGAPLARLEAAVALTALTSRFPGAQLA-GEPS
Mvan_5579|M.vanbaalenii_PYR-1 DRPVI-RHLGFGKGAHFCLGAPLARLEATLALTALTSRFPDAKLA-AEPV
MSMEG_6312|M.smegmatis_MC2_155 DRAQI-RHLGFGKGAHFCLGAPLARLEATVALPALAARFPEARLS-GEPE
MMAR_5268|M.marinum_M DRGAL-RHLGFGRGAHYCLGAPLARLEASVALSAIAARFPQARLD-GEPQ
MAV_0358|M.avium_104 DRKSL-RHLGFGRGVHYCLGAPLARLEAGVALSAVTARFPRARLD-GEPQ
MLBr_02088|M.leprae_Br4923 DRPSS-RHLAFAVGSHFCLGAALARLEATVTLSAISARFPQVQLA-GELV
MAB_0276|M.abscessus_ATCC_1997 DRSEI-KHLAFGHGPHFCLGAPLARLEARLALSSITARFPGARLM-DEPT
TH_1650|M.thermoresistible__bu GARTV-RDVGR-------------RSDRRVRP----ARAAAAHRT-GDGR
MUL_0475|M.ulcerans_Agy99 GREIGSKLLSFGSGAHFCLGAHLARMEARVALTELFKRIRGYQVDEGNAV
: :. * : : * :
Mb1912c|M.bovis_AF2122/97 RRDTRVLRGWSTLPVTLGPARSMVSP
Rv1880c|M.tuberculosis_H37Rv RRDTRVLRGWSTLPVTLGPARSMVSP
Mflv_1229|M.gilvum_PYR-GCK YKQNVTLRGMSELPITL---------
Mvan_5579|M.vanbaalenii_PYR-1 YKPNVTLRGMSELSITLA--------
MSMEG_6312|M.smegmatis_MC2_155 YKRNLTLRGMSTLSIAV---------
MMAR_5268|M.marinum_M YKTNVTLRGLESMTVALS--------
MAV_0358|M.avium_104 YKTNVTLRGLSRLVVAV---------
MLBr_02088|M.leprae_Br4923 YKPNVAMRGMSALPVQV---------
MAB_0276|M.abscessus_ATCC_1997 YKPNVTLRGLAELPVSIA--------
TH_1650|M.thermoresistible__bu VRTGPRAR------------------
MUL_0475|M.ulcerans_Agy99 RVHSSNVRGFAHLPITVEVA------
*