For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IAESRAEYGGVVTRVLSVPGGGPPIVLLHGYSDSALTWRPVLARLASAGRAAIAVDLPGFGHAAPRPPGP LLAQFDVFADAVLAAAGPAVLVGNSLGAATAVRAAARRENEVKALIALDDPLNSRHLIARIVRRWSVPEV FWAWVSRMPVPAGTLRRAARYFVPKMLYGPGVAADPEVVAYWCELISGPSAVARLGRDACRYAHEALGSH SGIEVSCPTLVVHGGRDRIIPIHSSGALQQQIPGSELVILPASGHCPQLDNPDEVTRLIVDFCDRISEPR QPDAAHSG*
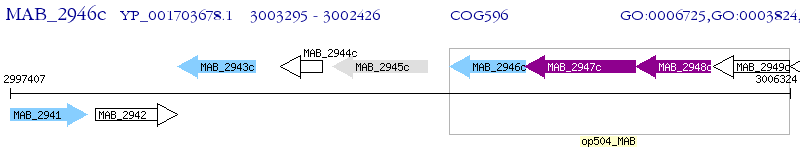
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2946c | - | - | 100% (289) | Alpha/beta hydrolase fold |
| M. abscessus ATCC 19977 | MAB_3685 | - | 5e-14 | 27.86% (280) | hypothetical protein MAB_3685 |
| M. abscessus ATCC 19977 | MAB_3810 | - | 3e-11 | 23.67% (283) | putative hydrolase, alpha/beta fold |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3196c | hpx | 7e-14 | 29.00% (269) | non-Heme haloperoxidase Hpx |
| M. gilvum PYR-GCK | Mflv_2414 | - | 2e-15 | 28.99% (276) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv3171c | hpx | 6e-14 | 29.00% (269) | non-Heme haloperoxidase Hpx |
| M. leprae Br4923 | MLBr_01921 | - | 4e-07 | 34.82% (112) | hypothetical protein MLBr_01921 |
| M. marinum M | MMAR_2050 | - | 5e-13 | 27.30% (282) | hypothetical protein MMAR_2050 |
| M. avium 104 | MAV_4061 | - | 1e-14 | 32.97% (276) | hydrolase, alpha/beta fold family protein |
| M. smegmatis MC2 155 | MSMEG_2517 | - | 4e-13 | 27.01% (274) | hydrolase, alpha/beta fold family protein, putative |
| M. thermoresistible (build 8) | TH_0684 | hpx | 1e-14 | 31.72% (268) | POSSIBLE NON-HEME HALOPEROXIDASE HPX |
| M. ulcerans Agy99 | MUL_2488 | hpx | 5e-13 | 29.35% (293) | non-heme haloperoxidase Hpx |
| M. vanbaalenii PYR-1 | Mvan_2343 | - | 4e-86 | 56.68% (277) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2946c|M.abscessus_ATCC_199 -----------------------------------------------MIA
Mvan_2343|M.vanbaalenii_PYR-1 -----------------------------------------------MIV
Mb3196c|M.bovis_AF2122/97 -------------------------------------------------M
Rv3171c|M.tuberculosis_H37Rv -------------------------------------------------M
MUL_2488|M.ulcerans_Agy99 ---------------------MSARKATDGKAHTKASAALPDVLPPSRSL
MAV_4061|M.avium_104 ---------------------MRAR-----------TDPFPDRLPPSRTV
TH_0684|M.thermoresistible__bu ----------------------------------------VEQMPPGRVV
MMAR_2050|M.marinum_M MRPTLAAAALAGASGLVGWGGARALARRIERNPDPFPRERLLAEPQGERV
Mflv_2414|M.gilvum_PYR-GCK ---------------------------------------------MGVTF
MSMEG_2517|M.smegmatis_MC2_155 ----------------------------------------------MIRS
MLBr_01921|M.leprae_Br4923 --------------------------------------------------
MAB_2946c|M.abscessus_ATCC_199 ESRAEYGGVVTRVLSVPGGGPPIVLLHG---YSDSALTWRPVLARLASAG
Mvan_2343|M.vanbaalenii_PYR-1 ETRASFGGVGTRVLAVPGSDPPVVLLHG---YADSADTWTEVLRGFGAAG
Mb3196c|M.bovis_AF2122/97 TVRAADGTPLHTQVFGPPHGYPIVLTHG---FVCAIRAWAYQIADLAGDY
Rv3171c|M.tuberculosis_H37Rv TVRAADGTPLHTQVFGPPHGYPIVLTHG---FVCAIRAWAYQIADLAGDY
MUL_2488|M.ulcerans_Agy99 TVRAADGTALHTQVFGPADGYPIVLTHG---FVCSIRAWAYQIADLATDY
MAV_4061|M.avium_104 TVRAVDGTRLHAQVFGPPDGYPIVLTHG---ITCAIRAWAYQIDDLARDY
TH_0684|M.thermoresistible__bu SVTAPDGVRLHAEVFGPADAPPIVLAHG---ITCAIRVWAHQIAELADDY
MMAR_2050|M.marinum_M AITRPDGTVLHALVAG--RGTPVILVHG---YCASLTEWNFVWDELLARG
Mflv_2414|M.gilvum_PYR-GCK ERNTVLVEGLTTSYLEAGQGDPIVLLHGGEFGADAALGWEATIDALASRY
MSMEG_2517|M.smegmatis_MC2_155 EVDLSGGRVSYLTWSPERPSGTVVLLHGGG-VDNAELSWGGIAPGLAAAG
MLBr_01921|M.leprae_Br4923 -------MSTNLLTLRGGRGKPLILVHG---LMGRGNTWSRQLPWLSQLG
.::* ** * :
MAB_2946c|M.abscessus_ATCC_199 RAAIAVDLPGFGHAAPRPP------GPLLAQFDVFADAVLAAAGPAVLVG
Mvan_2343|M.vanbaalenii_PYR-1 RRAVAVDLPGFGRAGARAP------GPLLGQFDAFAGALLKDLGPVVLVG
Mb3196c|M.bovis_AF2122/97 R-VIAFDHRGHGRSGVPRR-GAYSLNHLAADLDSVLDATLAPRERAVVAG
Rv3171c|M.tuberculosis_H37Rv R-VIAFDHRGHGRSGVPRR-GAYSLNHLAADLDSVLDATLAPRERAVVAG
MUL_2488|M.ulcerans_Agy99 R-VIAFDHRGHGRSGIPRR-GGYTLEHLASDLDSVLDATLAPHERALIAG
MAV_4061|M.avium_104 R-VIAYDHRGHGRSGVPKA-TGYSLKHLAADLDSVLEATLAPHERAVLAG
TH_0684|M.thermoresistible__bu R-VIAFDHRGHGRSSVPRRRWQYSLDHLASDLDAVLDATLDPGERAVIAG
MMAR_2050|M.marinum_M LRVIAFDQRGHGRSTLGAE--GIGSEPMADDLAAVLQHFDIRDG--VLVG
Mflv_2414|M.gilvum_PYR-GCK R-VLAPDMLGFGGSAKVVD---FTDGRGMRIRHIARFCDVMGIAAAHFVG
MSMEG_2517|M.smegmatis_MC2_155 YRVVCPDHPGYGHSPLPAL--PVTQERLVAYVGDFVDAVV--DGRYAIGG
MLBr_01921|M.leprae_Br4923 T-VYTYDAPWHRGRNVVDP----YPISTERFVTDLADAVSTLGVPARLVG
. * . . *
MAB_2946c|M.abscessus_ATCC_199 NSLGAATAVRAAARRENEVKALIALDDPLN-SRHLIARIVRRWSVP----
Mvan_2343|M.vanbaalenii_PYR-1 NSLGAATAVRAANRHPALVAGLVALDDPVN-ARHWLARLARLRPVP----
Mb3196c|M.bovis_AF2122/97 HSMGGITIAAWSDRYRHKVRRRTDAVALINTTTGDLVRKVKLLSVPRELS
Rv3171c|M.tuberculosis_H37Rv HSMGGITIAAWSDRYRHKVRRRTDAVALINTTTGDLVRKVKLLSVPRELS
MUL_2488|M.ulcerans_Agy99 HSMGGIAISAWSERYRHKVHRRADAVALINTATGDLLRKIRMLSVPRELS
MAV_4061|M.avium_104 HSMGGITIAAWSNRYRHKVSRRADAVALINTTSGELIRQVQLLSVPPGLS
TH_0684|M.thermoresistible__bu HSMGGIAISSWAERYPERVSERAAAVALINTTTGDLLHNVQLLPVPDRFA
MMAR_2050|M.marinum_M HSMGGFVSIRALLDHGALAQRLRAVVLFATWAGRVLDGAPQ---------
Mflv_2414|M.gilvum_PYR-GCK NSMGAINLLVDATSDTP----VLPMRSLVAVCGGGDIQRNEHVSALYDYD
MSMEG_2517|M.smegmatis_MC2_155 LSLGGGMTIGHVLDRPDRVTGAMLLGSYGIMARLSDGPFSAIRQALT---
MLBr_01921|M.leprae_Br4923 HSMGAMHSWCLAAERPDLVSALVVEDMAPDFRGCSTGPWEP---------
*:*.
MAB_2946c|M.abscessus_ATCC_199 -EVFWAWVSRMPVPAGTLRRAARYFVPKMLYG---PGVAADPEVVAYWCE
Mvan_2343|M.vanbaalenii_PYR-1 -APVWRGVGAIRLPAPAARALTEAAARKVLYG---PDAVVNPDVVSRWSS
Mb3196c|M.bovis_AF2122/97 PVRVLAGRSLVNTFGGFPLPGAARALSRHVISTLAVAADADPSATRLVYE
Rv3171c|M.tuberculosis_H37Rv PVRVLAGRSLVNTFGGFPLPGAARALSRHVISTLAVAADADPSATRLVYE
MUL_2488|M.ulcerans_Agy99 AARDFAGRGLINAFGGFPLPSAVRIPTQYLIAMMAVGADAHPTAARLLYE
MAV_4061|M.avium_104 PARVLAGRVLIGAFAGFAVPRAARVPSRHLVALLATGRDADPAIARLVYE
TH_0684|M.thermoresistible__bu AVRVRTASAVIKTVAGMPARLAA-VPSRYVVSMMAVGRDADPAITNFVHE
MMAR_2050|M.marinum_M -----NRLQIPLLERGILQRLLRSRTIAVLFGAAQCGARPSPAMISVFLE
Mflv_2414|M.gilvum_PYR-GCK ATLEGMRRIVTALFADPSYPADDEYVRRRYESSIAPGAWESLAAARFRRP
MSMEG_2517|M.smegmatis_MC2_155 WAMVRTGVMGSVMRWTARNRTAMNQSVAAIIRNRVYRTDDLLQAVYAAAA
MLBr_01921|M.leprae_Br4923 ----WLRGIPVEFDSVEKVFDEFGPVAGQYFLDAFDRTSTGWQLHGHTSR
MAB_2946c|M.abscessus_ATCC_199 LISGPSAVARLGRDACRYAHEALGSHS-GIEVSCPTLVVHGGRDRIIPIH
Mvan_2343|M.vanbaalenii_PYR-1 LVTQPRALETLGRYAFQYAIETAGGHR-EVRVNCPTLIVHGGRDRIIPVH
Mb3196c|M.bovis_AF2122/97 LFTQMSAAGRGGCAKMLVEEVGSAHLN-LDGLTVPTLVIGGVRDRLTPIS
Rv3171c|M.tuberculosis_H37Rv LFTQTSAAGRGGCAKMLVEEVGSAHLN-LDGLTVPTLVIGGVRDRLTPIS
MUL_2488|M.ulcerans_Agy99 LFAQTSAAGRGGCARMLVGEVGSRHIS-LDGLTVPTLVIGSERDRLTPIS
MAV_4061|M.avium_104 LFENTAPAGRAGCARALVSALGSRYLD-LAGLTVPTLVIGSERDRLTPIR
TH_0684|M.thermoresistible__bu LFTETPPAARDGWARTLVDAMGPRHIG-LHNLTVPTLVIGSDRDRLLPLV
MMAR_2050|M.marinum_M WFHQHLDRHGPHLPIVRAFSREDRYPR-LAEISVPAVVMVGSSDRTTPPS
Mflv_2414|M.gilvum_PYR-GCK GLEPPPTP--------------SSSRA-YERITVPTLVVEGGADKLLPRG
MSMEG_2517|M.smegmatis_MC2_155 QPGGFIAFEQWQRDQVRWNRQRTDYTDRLRTFPRPALIVHGGRDTGVPVA
MLBr_01921|M.leprae_Br4923 WIEIAAEWG------------TRDYWEHWLAVRSPVLLIEAG-TSVTPHG
. *.::: . *
MAB_2946c|M.abscessus_ATCC_199 SSGALQQQIPGSELVILPASGHCPQLDNPDEVTRLIVDFCDRISEPRQPD
Mvan_2343|M.vanbaalenii_PYR-1 ASRILHRQIPGSDLVVLPRSGHCPQLDDPAAVVRLTLRLLDGIPR-----
Mb3196c|M.bovis_AF2122/97 QSRRIARTAPNVVGLVELPGGHCSMLERHQEVNSHLRALAESVTRHVRDR
Rv3171c|M.tuberculosis_H37Rv QSRRIARTAPNVVGLVELPGGHCSMLERHQEVNSHLRALAESVTRHVRDR
MUL_2488|M.ulcerans_Agy99 QTRKIASTAPNVFDLVELPGGHCSMLEQHHEVNRHLRTLVDSVGGRSRAR
MAV_4061|M.avium_104 QSRRIAEGLPNLIELVELPGGHCSMLEHPREVNRRLRALAESATRNLPTH
TH_0684|M.thermoresistible__bu SSRRIAEAAPNLFDFVELPGGHCAILECPDRVNDHLRALAAAAAD---AR
MMAR_2050|M.marinum_M HSHRLAAGIPGARLVTVPGAGHLLNWEAVDELAEVVESFAAQR-------
Mflv_2414|M.gilvum_PYR-GCK WAAEIAGQIAGARSAVIDEAGHCPQIEQPEAVIGLLLTFFEEQR------
MSMEG_2517|M.smegmatis_MC2_155 HARAAARLIPDATLKVVAEAGHWVQRDAPDVVLDAMTTFLARISATA---
MLBr_01921|M.leprae_Br4923 QMLAMTKTACRTTYLYVPEAGHLIHDEATQVYRQAVVSFLAGCAGRD---
.** : :
MAB_2946c|M.abscessus_ATCC_199 AAHSG
Mvan_2343|M.vanbaalenii_PYR-1 -----
Mb3196c|M.bovis_AF2122/97 RISS-
Rv3171c|M.tuberculosis_H37Rv RISS-
MUL_2488|M.ulcerans_Agy99 -ISS-
MAV_4061|M.avium_104 RISS-
TH_0684|M.thermoresistible__bu RATS-
MMAR_2050|M.marinum_M -----
Mflv_2414|M.gilvum_PYR-GCK -----
MSMEG_2517|M.smegmatis_MC2_155 -----
MLBr_01921|M.leprae_Br4923 -----