For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRPTLAAAALAGASGLVGWGGARALARRIERNPDPFPRERLLAEPQGERVAITRPDGTVLHALVAGRGTP VILVHGYCASLTEWNFVWDELLARGLRVIAFDQRGHGRSTLGAEGIGSEPMADDLAAVLQHFDIRDGVLV GHSMGGFVSIRALLDHGALAQRLRAVVLFATWAGRVLDGAPQNRLQIPLLERGILQRLLRSRTIAVLFGA AQCGARPSPAMISVFLEWFHQHLDRHGPHLPIVRAFSREDRYPRLAEISVPAVVMVGSSDRTTPPSHSHR LAAGIPGARLVTVPGAGHLLNWEAVDELAEVVESFAAQR
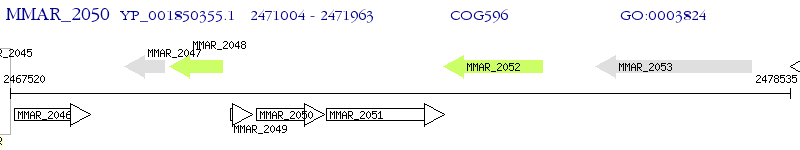
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_2050 | - | - | 100% (319) | hypothetical protein MMAR_2050 |
| M. marinum M | MMAR_1120 | - | 4e-17 | 27.83% (327) | hydrolase |
| M. marinum M | MMAR_1388 | hpx | 1e-14 | 30.32% (277) | non-heme haloperoxidase Hpx |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3196c | hpx | 4e-15 | 30.17% (295) | non-Heme haloperoxidase Hpx |
| M. gilvum PYR-GCK | Mflv_3146 | - | 2e-21 | 30.62% (258) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv3171c | hpx | 4e-15 | 30.17% (295) | non-Heme haloperoxidase Hpx |
| M. leprae Br4923 | MLBr_02269 | - | 7e-13 | 27.73% (238) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_3738c | - | 5e-20 | 30.77% (338) | putative hydrolase alpha/beta fold |
| M. avium 104 | MAV_4061 | - | 1e-21 | 33.79% (293) | hydrolase, alpha/beta fold family protein |
| M. smegmatis MC2 155 | MSMEG_4707 | - | 2e-24 | 31.66% (259) | non-heme bromoperoxidase BPO-A2 |
| M. thermoresistible (build 8) | TH_3727 | - | 2e-24 | 32.55% (255) | PUTATIVE Chloride peroxidase |
| M. ulcerans Agy99 | MUL_3299 | - | 1e-125 | 99.55% (223) | hypothetical protein MUL_3299 |
| M. vanbaalenii PYR-1 | Mvan_1567 | - | 4e-22 | 33.77% (231) | chloride peroxidase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2050|M.marinum_M --MRPTLAAAALAGASGLVGWGGA-RALARRIERNPDPFPRERLLAEPQG
MUL_3299|M.ulcerans_Agy99 --MRPTLAAAALAGASGLVGWGGA-RALARRIERNPDPFPRERLLAEPQG
Mb3196c|M.bovis_AF2122/97 --------------------------------------------------
Rv3171c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_4061|M.avium_104 -------------------------------MRARTDPFPDR----LPPS
Mflv_3146|M.gilvum_PYR-GCK --------------------------------------------------
TH_3727|M.thermoresistible__bu --------------------------------------------------
Mvan_1567|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_4707|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3738c|M.abscessus_ATCC_199 MSNKSNAWLAGVAGLGAVVAVAGVGTARSIGRRRFDDPYRGENFDLLQTD
MLBr_02269|M.leprae_Br4923 --------------------------------------------------
MMAR_2050|M.marinum_M ERVAITRPDG----TVLHALVAG---RGTPVILVHGYCASLTEWN-FVWD
MUL_3299|M.ulcerans_Agy99 ERVAITRPDG----TVLHALVAG---RGTPVILVHGYCASLTEWN-FVWD
Mb3196c|M.bovis_AF2122/97 --MTVRAADG----TPLHTQVFGPP-HGYPIVLTHGFVCAIRAWA-YQIA
Rv3171c|M.tuberculosis_H37Rv --MTVRAADG----TPLHTQVFGPP-HGYPIVLTHGFVCAIRAWA-YQIA
MAV_4061|M.avium_104 RTVTVRAVDG----TRLHAQVFGPP-DGYPIVLTHGITCAIRAWA-YQID
Mflv_3146|M.gilvum_PYR-GCK -MPTITTQDG----VEIFYKDWG---SGQPIVFSHGWPLSADDWD-AQLM
TH_3727|M.thermoresistible__bu -MATITTSDG----VEIFYRDWG---SGQPIVFSHGWPLTADDWD-PQML
Mvan_1567|M.vanbaalenii_PYR-1 -MSIFTTTDG----TDIFYKDWG---SGQPIVFSHGWPLSSDDWD-TQML
MSMEG_4707|M.smegmatis_MC2_155 -MAAITAYRGLLRSIELHVEDTGG--AGRPVVLVHGWPLTGESWG-AQVE
MAB_3738c|M.abscessus_ATCC_199 RGSIVTTDDG----VPLAVREVGPSNAPLTVVFVHGFCLQMASFH-FQRR
MLBr_02269|M.leprae_Br4923 -------------MINLAYEDRG---TGEPVVFIAGRGGAGRTWQPHQVP
: * .::: * :
MMAR_2050|M.marinum_M ELLAR---GLRVIAFDQRGHGRSTLG-AEGIGSEPMADDLAAVLQHFDIR
MUL_3299|M.ulcerans_Agy99 ELLAR---GLRVIAFDQRGHGRSTLG-AEGIGSEPMADDLAAVLQHFDLR
Mb3196c|M.bovis_AF2122/97 DLAG----DYRVIAFDHRGHGRSGVPRRGAYSLNHLAADLDSVLDATLAP
Rv3171c|M.tuberculosis_H37Rv DLAG----DYRVIAFDHRGHGRSGVPRRGAYSLNHLAADLDSVLDATLAP
MAV_4061|M.avium_104 DLAR----DYRVIAYDHRGHGRSGVPKATGYSLKHLAADLDSVLEATLAP
Mflv_3146|M.gilvum_PYR-GCK FFLGH---GYRVVAHDRRGHGRSGQV-DDGHDMDHYADDLAAVVEHLDLH
TH_3727|M.thermoresistible__bu FFLNA---GYRVIAHDRRGHGRSSPV-PDGHDMDHYADDLAALVEHLGLR
Mvan_1567|M.vanbaalenii_PYR-1 FFLQR---GYRVIAHDRRGHGRSTQT-PDGHDLDHYADDLAELTAHLDLR
MSMEG_4707|M.smegmatis_MC2_155 ALTAA---GHRVITYDRRGFGRSDTS-LVGYTYEALSDDLAAVLDELDVR
MAB_3738c|M.abscessus_ATCC_199 ELASRWGDNVRMVFYDQRGHGRSGLPAPASCTVGQLGDDLESVLRVLVPR
MLBr_02269|M.leprae_Br4923 AFLAA---GYRVITFDNRGIGATENT--EGFTTQTMVADTAVLIESLGAV
: . *:: .*.** * : . * :
MMAR_2050|M.marinum_M DG--VLVGHSMGGFVSIRALLDHG--ALAQRLRAVVLFATWAGRVLDGAP
MUL_3299|M.ulcerans_Agy99 DG--VLVGHSMGGFVSIRALLDHG--ALAQRLRAVVLFATWAGRVLDGAP
Mb3196c|M.bovis_AF2122/97 RERAVVAGHSMGG-ITIAAWSDRYRHKVRRRTDAVALINTTTGDLVRKVK
Rv3171c|M.tuberculosis_H37Rv RERAVVAGHSMGG-ITIAAWSDRYRHKVRRRTDAVALINTTTGDLVRKVK
MAV_4061|M.avium_104 HERAVLAGHSMGG-ITIAAWSNRYRHKVSRRADAVALINTTSGELIRQVQ
Mflv_3146|M.gilvum_PYR-GCK DA--VHVGHSTGGGEVVRYLARHG----EDRAVKAALISAVPPLMVQTDT
TH_3727|M.thermoresistible__bu DA--VHVGHSTGGGVVVRYLARHG----EERTAKAVLIAAVPPLMVRTDA
Mvan_1567|M.vanbaalenii_PYR-1 DA--IHVGHSTGGGEVVRYLGRHG----ESRVAKAALISAVPPLMVRTED
MSMEG_4707|M.smegmatis_MC2_155 DA--TLVGFSMGTGEVAAYCARKG----TERLRSVVLAAPVTPYMLNTAD
MAB_3738c|M.abscessus_ATCC_199 GN-AVLVGHSMGGMTVLAHARRHP-EQYGRRIVGVGLIASAAEGLSHTAI
MLBr_02269|M.leprae_Br4923 PAR--IVGVSMGSFIAQELMVARP------ELVRAAVLMATRGRLDRTR-
.* * * : . . : . :
MMAR_2050|M.marinum_M QNRLQIPLLERGILQR--LLRSRTIAVLFGAAQCGARPSPAMISVF----
MUL_3299|M.ulcerans_Agy99 QNRLQIPLLERGILQR--LLRSRTIAVLFGAAQCGARPSPAMISG-----
Mb3196c|M.bovis_AF2122/97 LLSVPRELSPVRVLAGRSLVNTFGGFPLPGAARALSRHVISTLAVAADAD
Rv3171c|M.tuberculosis_H37Rv LLSVPRELSPVRVLAGRSLVNTFGGFPLPGAARALSRHVISTLAVAADAD
MAV_4061|M.avium_104 LLSVPPGLSPARVLAGRVLIGAFAGFAVPRAARVPSRHLVALLATGRDAD
Mflv_3146|M.gilvum_PYR-GCK NPGG-LPKSVFDDFQHQVATNRAGFYRAVPEGPFYGFNREGVEPV-----
TH_3727|M.thermoresistible__bu NPGG-LPKSVFDDFQAQLAANRSEFYRALASGPFYGFNRDGVEPS-----
Mvan_1567|M.vanbaalenii_PYR-1 NPEG-TPKEVFDDLQAQLAANRSVFYRSLPAGPFYGFNRPGVEPQ-----
MSMEG_4707|M.smegmatis_MC2_155 NPEGPLSKTQAAQMAAALTTNSDAFYREMMTG---VFSADGVLTIS----
MAB_3738c|M.abscessus_ATCC_199 GEGLRNPALRVLRTAVHYAPGPAHHGRGAIKSLVGPVLQAASYGGHR---
MLBr_02269|M.leprae_Br4923 -QFFHAAEAEFHDSGIQLPSGYNAKVRLLENLSRKTLNDDVAVAD-----
MMAR_2050|M.marinum_M --LEWFHQHLDRHGPHLPIVRA--FSREDRYPRLAEISVP---AVVMVGS
MUL_3299|M.ulcerans_Agy99 --IPRVVPSTSRPARTAPTHRAGVLARGSLSTAGRDLGAGG-GDGGLLGS
Mb3196c|M.bovis_AF2122/97 PSATRLVYELFTQMSAAGRGGCAKMLVEEVGSAHLNLDGLT-VPTLVIGG
Rv3171c|M.tuberculosis_H37Rv PSATRLVYELFTQTSAAGRGGCAKMLVEEVGSAHLNLDGLT-VPTLVIGG
MAV_4061|M.avium_104 PAIARLVYELFENTAPAGRAGCARALVSALGSRYLDLAGLT-VPTLVIGS
Mflv_3146|M.gilvum_PYR-GCK --EGIIANWWRQGMQGGAKAHYDGIVAFSQTDFTEDLQKIR-IPVLVMHG
TH_3727|M.thermoresistible__bu --EALIANWWRQGMMGDAKAHYDGIVAFSQTDFTEDLTKIT-VPVLVMHG
Mvan_1567|M.vanbaalenii_PYR-1 --EAIIENWWRQGMMGDAYSHYDGIVAFSQTDFTEDLKKIT-VPVLVMHS
MSMEG_4707|M.smegmatis_MC2_155 --EDALQQVLDMCGQASKNAALACMAAFSNTDFREDLPRVT-VPALVIHG
MAB_3738c|M.abscessus_ATCC_199 VSPTLVKFSERMIHQTPVTTIVDFLRALEQHDETAALPTIAPLPSLVICG
MLBr_02269|M.leprae_Br4923 ----WIAMFNMWPIKSTPGLRCQTDVAPQNNRLPAYRSIAA--PVLVIGF
. ::
MMAR_2050|M.marinum_M -SDRTTPPSHSHRLAAGIPG---ARLVT--VPGAGHLLN--WEAVDELAE
MUL_3299|M.ulcerans_Agy99 -HHPAQPLAPAGRRNPGCPAGHRARRGTPAELGGGRRAGRSRRILCGAAV
Mb3196c|M.bovis_AF2122/97 VRDRLTPISQSRRIARTAPNVVGLVELPGGHCSMLERHQEVNSHLRALAE
Rv3171c|M.tuberculosis_H37Rv VRDRLTPISQSRRIARTAPNVVGLVELPGGHCSMLERHQEVNSHLRALAE
MAV_4061|M.avium_104 ERDRLTPIRQSRRIAEGLPNLIELVELPGGHCSMLEHPREVNRRLRALAE
Mflv_3146|M.gilvum_PYR-GCK DDDQIVPYDDAGPLSAALLPNGILKTYNGFPHGMP--TTHADVINADLLE
TH_3727|M.thermoresistible__bu DDDQVVPYANSGPLTAELLRNAKLKTYQGFPHGMM--TTHAEVVNADILE
Mvan_1567|M.vanbaalenii_PYR-1 EADQIVPYVASGPKSAALLKNGVLKTYKDFPHGMP--TTHADVVNADLLE
MSMEG_4707|M.smegmatis_MC2_155 DADATTPLQNCGERTHAALPNSRLHVISGGPHGIP--VSHTEEFNRVLLD
MAB_3738c|M.abscessus_ATCC_199 DTDMLTPHTQSESMAARLAASGHNELILVRESGHLVQLEHPEIVNDGIDR
MLBr_02269|M.leprae_Br4923 AEDVVTPPSLGREVADVLPNGRYLQIPDAGHLGFFERPEAVNAAALQFFA
. * .
MMAR_2050|M.marinum_M VVESFAAQR--------------
MUL_3299|M.ulcerans_Agy99 LMRGHAAGVWR------------
Mb3196c|M.bovis_AF2122/97 SVTRHVRDRRISS----------
Rv3171c|M.tuberculosis_H37Rv SVTRHVRDRRISS----------
MAV_4061|M.avium_104 SATRNLPTHRISS----------
Mflv_3146|M.gilvum_PYR-GCK FIRS-------------------
TH_3727|M.thermoresistible__bu FLRS-------------------
Mvan_1567|M.vanbaalenii_PYR-1 FLKG-------------------
MSMEG_4707|M.smegmatis_MC2_155 FLAEDLAAAR-------------
MAB_3738c|M.abscessus_ATCC_199 LVRRSTPPLFAAIKQRLRDRTGL
MLBr_02269|M.leprae_Br4923 GVKGEWP----------------