For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
QLSQIAQKSGVAAAGFVVAGMLAATPALADPEDLDFGQKAEIPSPGGAIDYTVSTLEPSGHNDGVWYSDV TARAVSGSPTPNIADFNARAVNSSTYAVMKGDKPDGLPNQPLTAGSQTTGRIYFDVRGGTAPDSVVYRDA GGTDKVVWKD*
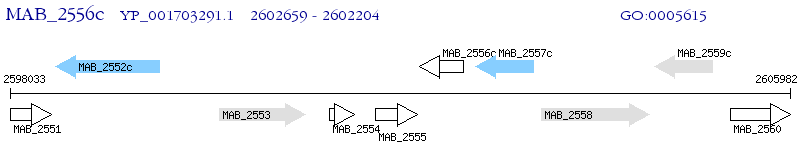
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2556c | - | - | 100% (151) | hypothetical protein MAB_2556c |
| M. abscessus ATCC 19977 | MAB_1026c | - | 1e-12 | 32.41% (145) | hypothetical protein MAB_1026c |
| M. abscessus ATCC 19977 | MAB_0462c | - | 1e-10 | 31.08% (148) | hypothetical protein MAB_0462c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1961c | mpt63 | 1e-10 | 34.88% (86) | immunogenic protein MPT63 (antigen MPT63/MPB63) (16 KD |
| M. gilvum PYR-GCK | Mflv_1515 | - | 2e-15 | 38.78% (147) | FHA domain-containing protein |
| M. tuberculosis H37Rv | Rv1926c | mpt63 | 1e-10 | 34.88% (86) | immunogenic protein MPT63 (antigen MPT63/MPB63) (16 kDa |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_1823 | - | 3e-16 | 37.33% (150) | hypothetical protein MMAR_1823 |
| M. avium 104 | MAV_3737 | - | 4e-19 | 35.03% (157) | hypothetical protein MAV_3737 |
| M. smegmatis MC2 155 | MSMEG_5617 | - | 5e-17 | 36.91% (149) | immunogenic protein MPT63 |
| M. thermoresistible (build 8) | TH_4151 | - | 1e-15 | 37.24% (145) | PUTATIVE putative FHA domain containing protein |
| M. ulcerans Agy99 | MUL_2071 | - | 8e-11 | 34.13% (126) | hypothetical protein MUL_2071 |
| M. vanbaalenii PYR-1 | Mvan_5241 | - | 2e-16 | 35.17% (145) | FHA domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1823|M.marinum_M -------MKTAVGAAAIAAASVFGAASASAAPPTIQGFGTSERLVDG--P
MUL_2071|M.ulcerans_Agy99 MEFIATTMKTAVGAAAIAAASVFGAASASAAPPTIQGFGTSERLVDG--P
MAV_3737|M.avium_104 MKVTKNAVLTAAAAGGIAAASVFAATPAVAAP-NIQGFGVSEQLVDG--P
Mb1961c|M.bovis_AF2122/97 -MKLTTMIKTAVAVVAMAAIATFAAPVALAAYPITGKLGSELTMTDTVGQ
Rv1926c|M.tuberculosis_H37Rv -MKLTTMIKTAVAVVAMAAIATFAAPVALAAYPITGKLGSELTMTDTVGQ
TH_4151|M.thermoresistible__bu ---MNSKILAAPVVLATAAGLALATAAPAPARPGVADFGATQHLHD--GG
Mflv_1515|M.gilvum_PYR-GCK -MKFKPALVAAATAAAATAVA-LGGAATASADTEVQWLGQPGELVNG-T-
Mvan_5241|M.vanbaalenii_PYR-1 -MKFKPTVAAIITTAAAAAVG-VVGAGSASADAEVQYLGQPGELVNG-S-
MSMEG_5617|M.smegmatis_MC2_155 -MNIRTGMKIAAASTATAAMA-LGAAPLAMAETEAG-FGEAQDLVAP-TG
MAB_2556c|M.abscessus_ATCC_199 -MQLSQIAQKSGVAAAGFVVAGMLAATPALADPEDLDFGQKAEIPSP--G
. . . . * :* :
MMAR_1823|M.marinum_M MVTSYTVGNLQLSDVTIAGYLPQGQLYSADVTARSVTGTITPLVSSFNAR
MUL_2071|M.ulcerans_Agy99 MVTSYTVGNLQLSDVTIAGYLPQGQLYSADVTGRSVTGTITPLVSSFSAR
MAV_3737|M.avium_104 LVTDYTVSNLQPSNVSIPGYTPKGTLYQADISVRSDGGVVTPMVHDFIAR
Mb1961c|M.bovis_AF2122/97 VVLGWKVSDLKSSTAVIPGYPVAGQVWEATATVNAIRGSVTPAVSQFNAR
Rv1926c|M.tuberculosis_H37Rv VVLGWKVSDLKSSTAVIPGYPVAGQVWEATATVNAIRGSVTPAVSQFNAR
TH_4151|M.thermoresistible__bu VTIAYTLEELEPSDDVVSNVALQGRLWEVSVTVEAVEGAATPVIPFFNAR
Mflv_1515|M.gilvum_PYR-GCK VVQHWTVSHLKPSTDTIP-YQPVGTLWEATASDEAVAGNVTPIISNLNAR
Mvan_5241|M.vanbaalenii_PYR-1 VIQHWTVTGLKPSSDVIP-YRPVGTLWEATATDEAIAGTVTPIVSNLNAR
MSMEG_5617|M.smegmatis_MC2_155 DTASVTVTDIALSSDQIP-TEVEGQLYEATATIEAKEGTVTPVLSNFNAR
MAB_2556c|M.abscessus_ATCC_199 GAIDYTVSTLEPSG-------HNDGVWYSDVTARAVSGSPTPNIADFNAR
.: : * . :: : .: * ** : : **
MMAR_1823|M.marinum_M AANGDTYGVVNEVPAPGGLNPAPILQGNSQSGVLYFDVTG-PPPNGVVYN
MUL_2071|M.ulcerans_Agy99 AANGDTYGVVNEVPAPGGLNPAPILQGNSQSGVLYF--------------
MAV_3737|M.avium_104 GPNGQNYRVIDNVAVPNGLSPAPIPQGTESTGTLYFDVTG-APPNGVVYN
Mb1961c|M.bovis_AF2122/97 TADGINYRVLWQAAGPDTISGATIPQGEQSTGKIYFDVTG-PSPTIVAMN
Rv1926c|M.tuberculosis_H37Rv TADGINYRVLWQAAGPDTISGATIPQGEQSTGKIYFDVTG-PSPTIVAMN
TH_4151|M.thermoresistible__bu SADGQNYRALFTAVGEEALSGATLGQGQETEGNIYFDVTG-APPTEVVYN
Mflv_1515|M.gilvum_PYR-GCK AKNGDTYRVLFGVATTQGVNPAPLAQGHKTSGKIYFDVTG-EAPDSVFYS
Mvan_5241|M.vanbaalenii_PYR-1 AENGDTYRVLFGVATEQGVNPSTLAQGQKTTGKIYFDVTG-QQPDSVFYS
MSMEG_5617|M.smegmatis_MC2_155 TADGQSYQAVYEVPASEG-NPEPLTEGASTTGKVYFDVKG-EAPDSVVYS
MAB_2556c|M.abscessus_ATCC_199 AVNSSTYAVMKGDK-PDGLPNQPLTAGSQTTGRIYFDVRGGTAPDSVVYR
:. .* .: .: * . * :**
MMAR_1823|M.marinum_M DGV-QDVLIWTSNV------------------------------------
MUL_2071|M.ulcerans_Agy99 --------------------------------------------------
MAV_3737|M.avium_104 DGM-QDVLMWTSNVPSSAPGAPNSPAPGAPNSPAPGAPPAPAAHT-----
Mb1961c|M.bovis_AF2122/97 NGM-EDLLIWEP--------------------------------------
Rv1926c|M.tuberculosis_H37Rv NGM-EDLLIWEP--------------------------------------
TH_4151|M.thermoresistible__bu DAV-QDRLIWR---------------------------------------
Mflv_1515|M.gilvum_PYR-GCK TGD-AEELLWLQAPPPATGSGTSGSTSGSSSSGASSAPSAAAETTAPETP
Mvan_5241|M.vanbaalenii_PYR-1 DGG-SEVLLWLEAPP-APATGSAG--SGASGYGSEASPSAAADTATPETP
MSMEG_5617|M.smegmatis_MC2_155 DVG-SDEVIWTDDPGQ----------------------------------
MAB_2556c|M.abscessus_ATCC_199 DAGGTDKVVWKD--------------------------------------
MMAR_1823|M.marinum_M --------------------------------------------------
MUL_2071|M.ulcerans_Agy99 --------------------------------------------------
MAV_3737|M.avium_104 --------------------------------------------------
Mb1961c|M.bovis_AF2122/97 --------------------------------------------------
Rv1926c|M.tuberculosis_H37Rv --------------------------------------------------
TH_4151|M.thermoresistible__bu --------------------------------------------------
Mflv_1515|M.gilvum_PYR-GCK ASTPAAEPAAVGSATPASVPSGSSGTPLPEGSSGTPLPEGSSGTPLPEGS
Mvan_5241|M.vanbaalenii_PYR-1 GTTPAAETPTSVVPTPVAAPAGSSGTPLPEGSSGTPLPEGSSGTPLPAGA
MSMEG_5617|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2556c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1823|M.marinum_M --------------------------------------------------
MUL_2071|M.ulcerans_Agy99 --------------------------------------------------
MAV_3737|M.avium_104 --------------------------------------------------
Mb1961c|M.bovis_AF2122/97 --------------------------------------------------
Rv1926c|M.tuberculosis_H37Rv --------------------------------------------------
TH_4151|M.thermoresistible__bu --------------------------------------------------
Mflv_1515|M.gilvum_PYR-GCK S-----GTPLPANAAPPAEAAP-APVPAAA--------TPAPAAPTTTVA
Mvan_5241|M.vanbaalenii_PYR-1 APVEAGPAPVPAGVAPAPHGSSGTPLPAGAPAVEGVQPAPAPGAPTTTVV
MSMEG_5617|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2556c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_1823|M.marinum_M ------
MUL_2071|M.ulcerans_Agy99 ------
MAV_3737|M.avium_104 ------
Mb1961c|M.bovis_AF2122/97 ------
Rv1926c|M.tuberculosis_H37Rv ------
TH_4151|M.thermoresistible__bu ------
Mflv_1515|M.gilvum_PYR-GCK VPAPPA
Mvan_5241|M.vanbaalenii_PYR-1 VPAPPA
MSMEG_5617|M.smegmatis_MC2_155 ------
MAB_2556c|M.abscessus_ATCC_199 ------