For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTTADGRALHYQVAGSGEPTVVFESGMGFSRTAWGLVQPLVARRLRSVVYDRANLGRSDDDPAPRTLERI TGDLDALLTALGSGPYILVGHSYGGTIALAATATEPSRIAGVVLVDHSDENLEAYYRPASPGMRLLGLAH RAALDILGRVGLLPHVVRRMMPRMPRDVLDDMVSEDLTPRARRAANEEDRRFVAGMTELRSNPPALDGVP VTVISGARAELLNEETRSELNAAHQMTARRLGARHVVARKSGHQVVLTEPRLIADEVFRIAGA
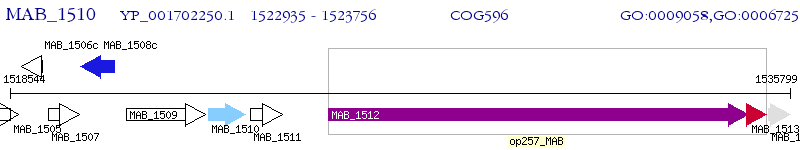
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1510 | - | - | 100% (273) | Alpha/beta hydrolase fold precursor |
| M. abscessus ATCC 19977 | MAB_0470c | - | 2e-79 | 57.84% (268) | putative hydrolase (alpha/beta hydrolase fold) |
| M. abscessus ATCC 19977 | MAB_3738c | - | 8e-11 | 28.62% (311) | putative hydrolase alpha/beta fold |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2734 | - | 7e-09 | 24.91% (289) | hydrolase |
| M. gilvum PYR-GCK | Mflv_4667 | - | 8e-10 | 36.79% (106) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv2715 | - | 7e-09 | 24.91% (289) | hydrolase |
| M. leprae Br4923 | MLBr_00862 | ephD | 3e-06 | 24.49% (294) | short chain dehydrogenase |
| M. marinum M | MMAR_5090 | - | 3e-09 | 30.71% (140) | hydrolase |
| M. avium 104 | MAV_2275 | - | 3e-10 | 27.52% (298) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_1587 | - | 1e-13 | 24.43% (307) | alpha/beta hydrolase fold |
| M. thermoresistible (build 8) | TH_0447 | lipV | 3e-09 | 34.86% (109) | POSSIBLE LIPASE LIPV |
| M. ulcerans Agy99 | MUL_4166 | - | 2e-09 | 30.71% (140) | hydrolase |
| M. vanbaalenii PYR-1 | Mvan_5882 | - | 1e-09 | 32.40% (179) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2734|M.bovis_AF2122/97 -----------------------MTERKRNLRPVRDVAPPTLQFRTVHGY
Rv2715|M.tuberculosis_H37Rv -----------------------MTERKRNLRPVRDVAPPTLQFRTVHGY
MLBr_00862|M.leprae_Br4923 -----------------------------MPATQSIHKVPHQFVESPDGV
MAV_2275|M.avium_104 -----------------------------MPAPQ---HSPQHFVHSADGT
Mflv_4667|M.gilvum_PYR-GCK -----------------------------------------------MTE
TH_0447|M.thermoresistible__bu --------------------------------------------------
MMAR_5090|M.marinum_M -----------------------------------------------MPT
MUL_4166|M.ulcerans_Agy99 -----------------------------------------------MPT
Mvan_5882|M.vanbaalenii_PYR-1 -----------------------------------------MPLVPTRAG
MAB_1510|M.abscessus_ATCC_1997 --------------------------------------------MTTADG
MSMEG_1587|M.smegmatis_MC2_155 MSRHHRRNHLRLSRIAAVACVAWVAALFCGPPSAIAVPEDVAGLLDIGGG
Mb2734|M.bovis_AF2122/97 RRAFRIAGS--GPAILLIHGIGDNSTTWNGVHAKLAQRFTVIAPDLLGHG
Rv2715|M.tuberculosis_H37Rv RRAFRIAGS--GPAILLIHGIGDNSTTWNGVHAKLAQRFTVIAPDLLGHG
MLBr_00862|M.leprae_Br4923 RLAIYQSGQPEGPAIVLVHGFPDSHVLWDGVVPLLAKRFRIVRYDNRGVG
MAV_2275|M.avium_104 RIAVYDEGNPEGPTVVLVHGFPDSHVLWDGVVPLLAERFRILRYDNRGVG
Mflv_4667|M.gilvum_PYR-GCK QLCTYRFGPAGPARVLAIHGLTGHGKRWESLFTRHLSDIPAVAPDLVGHG
TH_0447|M.thermoresistible__bu ---VYRYGPAGPPRLLAIHGLTGHGRRWQTLATRHLPEIAVAAPDLIGHG
MMAR_5090|M.marinum_M DLLTYHGGEGEP--LVLVHGLMGRGSTWSRQLPWLTRLGAVYTYDAPWHR
MUL_4166|M.ulcerans_Agy99 DLLTYHGGEGEP--LVLVHGLMGRGSTWSRQLPWLTRLGAVYTYDAPWHR
Mvan_5882|M.vanbaalenii_PYR-1 TLAYHVRGS--GPTVVLLPAALHDHHDFDAVSADLARHHRVVAVDWPGCG
MAB_1510|M.abscessus_ATCC_1997 RALHYQVAGSGEPTVVFESGMGFSRTAWGLVQPLVARRLRSVVYDRANLG
MSMEG_1587|M.smegmatis_MC2_155 RAIFLECRGSGSPTVVLMAGKGNGADDWLQVLAPGDPEQDAPGDNLP-FG
:: . : . :
Mb2734|M.bovis_AF2122/97 QSDKPR--ADYSVAAYANGMRDLLSVLDIER-VTIVGHSLGGGVAMQFAY
Rv2715|M.tuberculosis_H37Rv QSDKPR--ADYSVAAYANGMRDLLSVLDIER-VTIVGHSLGGGVAMQFAY
MLBr_00862|M.leprae_Br4923 QSSVPKPVSAYTMTRFADDFAAVIDELSPDRPVHVLAHDWGSVGVWEYLS
MAV_2275|M.avium_104 ASSAPKPVSAYRMDRFADDFAAVIGELSPGGPVHVLAHDWGSVGVWHYLK
Mflv_4667|M.gilvum_PYR-GCK RSSWDAP---WTIEAN-VAALTALLDAEAEGPVVVVGHSFGGAIALNLAA
TH_0447|M.thermoresistible__bu RSSWAAP---WTFEAN-VGALVALLEDQADGPVVVVGHSFGGAVAMHLAA
MMAR_5090|M.marinum_M GRDVDDP---YPISTERFVADLGEAVAELGGPARLVGHSMGALHSWCLAA
MUL_4166|M.ulcerans_Agy99 GRDVDDP---YPISTERFVADLGEAVAELGGPARLVGHSMGALHSWCLAA
Mvan_5882|M.vanbaalenii_PYR-1 DSPPAPDPHAVTAPVLADGLEDLVAGLGEEP-VVLIGNSVGGFAAARLAI
MAB_1510|M.abscessus_ATCC_1997 RSDDDPAP--RTLERITGDLDALLTALGSGP-YILVGHSYGGTIALAATA
MSMEG_1587|M.smegmatis_MC2_155 RGDLIHS----SEAVLPAIARFTRVCAYDRPDVRMGGSDISTPRAQPHSV
: . . .
Mb2734|M.bovis_AF2122/97 QFPQLVDRLILVSAGGVTKDVNIVFRLASLPMGSEAMALLRLP-------
Rv2715|M.tuberculosis_H37Rv QFPQLVDRLILVSAGGVTKDVNIVFRLASLPMGSEAMALLRLP-------
MLBr_00862|M.leprae_Br4923 RSGASDRVASFTSVSGPSQDQLVDYILSGLRRPWRPRTFARAVSQLFRLT
MAV_2275|M.avium_104 RPGANDRVATFTSVSGPSQDQLVDYIFSGLRAPWRPRAFARALGQALRLS
Mflv_4667|M.gilvum_PYR-GCK ARPDLIAGLVLLDPAVGLDGRWMREIADEMYTSPDYTDREEAR-------
TH_0447|M.thermoresistible__bu ARPDLVAGLVLLDPAIGLDGEWMREIAESMLASPDYPDREEAR-------
MMAR_5090|M.marinum_M ERPGLVSALVVEDMAPDFVGRTTGPWEPWLHALPVEFDSADQV-------
MUL_4166|M.ulcerans_Agy99 ERPGLVSALVVEDMAPDFVGRTTGPWEPWLHALPVEFDSADQV-------
Mvan_5882|M.vanbaalenii_PYR-1 RRPHRVAGLVLVNTGGFLPINPIMRLFCRTLGTPQVARR-----------
MAB_1510|M.abscessus_ATCC_1997 TEPSRIAGVVLVDHSDENLEAYYRPASPGMRLLGLAHRAALDILG-----
MSMEG_1587|M.smegmatis_MC2_155 DLDVDDLHALLTALGEPAPYVLVAHSYSGLIADLYARMHPETVG------
. .
Mb2734|M.bovis_AF2122/97 --LVLPAVQIAGRIVGKAIGTTSLGHDLPNVLRILDDLPEPTASAAFGRT
Rv2715|M.tuberculosis_H37Rv --LVLPAVQIAGRIVGKAIGTTSLGHDLPNVLRILDDLPEPTASAAFGRT
MLBr_00862|M.leprae_Br4923 YMVLFSIPVLVPMVLWIALSSATIRRTMVDNISTDQIHHSATLARDAAHS
MAV_2275|M.avium_104 YMILLSIPVLAPLALRLTLSIPALRRNAVDNIPIEQIHHSDRLAADAARS
Mflv_4667|M.gilvum_PYR-GCK ---------------QEKIGGSWGEVAEDELDRELDEHLVASAGGRVGWR
TH_0447|M.thermoresistible__bu ---------------TEKFTGSWADVEPAELDRDLDEHLIALPGGRFGWR
MMAR_5090|M.marinum_M ---------------FAEFGPLAGQYFLEAFDRTATGWRLHGHTSRWIQI
MUL_4166|M.ulcerans_Agy99 ---------------FAEFGPLAGQYFLEAFDHTATGWRLHGHTSRWIQI
Mvan_5882|M.vanbaalenii_PYR-1 ---------------ILPAFIRLYMQADNDLARAARDRAVTRAKTTSGVA
MAB_1510|M.abscessus_ATCC_1997 ---------------RVGLLPHVVRRMMPRMPRDVLDDMVSEDLTPRARR
MSMEG_1587|M.smegmatis_MC2_155 ---ALVMVDAVSEQMADVVNPAALANWDASNATTSDQVREGVLLIDAFGR
Mb2734|M.bovis_AF2122/97 LRAVVD--WRGQMVTMLDRCYLTEAIPVQIIWGTKDVVLPVRHAHMAHAA
Rv2715|M.tuberculosis_H37Rv LRAVVD--WRGQMVTMLDRCYLTEAIPVQIIWGTKDVVLPVRHAHMAHAA
MLBr_00862|M.leprae_Br4923 VKTYPANYFRAFSGRRRGKGVPVVNVPVQLIVNTMDRYVRP-YGYDATAR
MAV_2275|M.avium_104 VKTYPANYFRSFAIRK--QGVAVIDVPVQLIVNTEDRYVRP-YGYDHTPR
Mflv_4667|M.gilvum_PYR-GCK ISIPAMLCYWSELTRPAVIPRDGTPTTLVRATRTDPPYAGDELVAALDAG
TH_0447|M.thermoresistible__bu ICVPAMMAYWSELARDVVLPPSGIPTTLVRAAWTDPPYVTGELLDGLSQR
MMAR_5090|M.marinum_M AAEWGTRDYWAQWR------AVRQPALLIEAGNSVTP--PGQMLAMAEVD
MUL_4166|M.ulcerans_Agy99 AAEWGTRDYWAQWR------AVRQPALLIEAGNSVTP--PGQMLAMAEVD
Mvan_5882|M.vanbaalenii_PYR-1 MTAALWRSFATDAHDLR-RDADKLTAPALIVWGSRDTAIPLPIGRATHTA
MAB_1510|M.abscessus_ATCC_1997 AANEEDRRFVAGMTELRSNPPALDGVPVTVISGARAELLNEETRSELNAA
MSMEG_1587|M.smegmatis_MC2_155 IAAAPGLPAVPAVVLSADKPWRTDLLPESARQGEQVTFADWLVAQDRLAA
Mb2734|M.bovis_AF2122/97 MPGSQLEIFEGSGHFPFHDDPARFIDIVERFMDTTEPAEYDQAALRALLR
Rv2715|M.tuberculosis_H37Rv MPGSQLEIFEGSGHFPFHDDPARFIDIVERFMDTTEPAEYDQAALRALLR
MLBr_00862|M.leprae_Br4923 WVPRLWRRDIKAGHFSPMSHPQVMAAAVHDLADLVDGKQPSRALLRAQVG
MAV_2275|M.avium_104 FVPRLWRRDIRAGHFSPMSHPQVMAAAVHDFADMAEGKPASRALLRAQVG
Mflv_4667|M.gilvum_PYR-GCK LGPNFTLLEWDCDHMVPLARPTETARLIRDLLP-----------------
TH_0447|M.thermoresistible__bu LGADLTLVDLDCLHMVAQAKPAETAELIRRHLG-----------------
MMAR_5090|M.marinum_M RSTKYLHVP-EAGHLVHDEAQLAYRQAVEAFLAAERVR------------
MUL_4166|M.ulcerans_Agy99 RSTKYLHVP-EAGHLVHDEAQLAYRQAVEAFLAAERVR------------
Mvan_5882|M.vanbaalenii_PYR-1 LP-RARFETLPTGHVPFASAPQDFLAILRPFLTRTLGTTAGSPHQ-----
MAB_1510|M.abscessus_ATCC_1997 HQMTARRLG--ARHVVARKSGHQVVLTEPRLIADEVFRIAGA--------
MSMEG_1587|M.smegmatis_MC2_155 AIGAGHITETHSGHDIYLYSPALVVEAIRDVVAQVTRVHG----------
*
Mb2734|M.bovis_AF2122/97 R-----GGGEATVTGS----------------ADTRVAVLNAIGSNERSA
Rv2715|M.tuberculosis_H37Rv R-----GGGEATVTGS----------------ADTRVAVLNAIGSNERSA
MLBr_00862|M.leprae_Br4923 RSRDTFGDTLVSVTGAGSGIGRETALALARRGAEVVVSDIDEAAAKNTAA
MAV_2275|M.avium_104 RPRKAFGDTLVSVTGAGSGIGRATAFAFAREGAELIVSDIDEAAVKATAA
Mflv_4667|M.gilvum_PYR-GCK --------------------------------------------------
TH_0447|M.thermoresistible__bu --------------------------------------------------
MMAR_5090|M.marinum_M --------------------------------------------------
MUL_4166|M.ulcerans_Agy99 --------------------------------------------------
Mvan_5882|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_1510|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_1587|M.smegmatis_MC2_155 --------------------------------------------------
Mb2734|M.bovis_AF2122/97 T-------------------------------------------------
Rv2715|M.tuberculosis_H37Rv T-------------------------------------------------
MLBr_00862|M.leprae_Br4923 QIVASGGVAYAYALDVSDAAAVEAFAEQVSAKHGVPDIVVNNAGIGQAGR
MAV_2275|M.avium_104 EIAGRGGVAHAYVLDVSDAQAVEEFAERVSAAHGVPDIVVNNAGIGQAGG
Mflv_4667|M.gilvum_PYR-GCK --------------------------------------------------
TH_0447|M.thermoresistible__bu --------------------------------------------------
MMAR_5090|M.marinum_M --------------------------------------------------
MUL_4166|M.ulcerans_Agy99 --------------------------------------------------
Mvan_5882|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_1510|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_1587|M.smegmatis_MC2_155 --------------------------------------------------
Mb2734|M.bovis_AF2122/97 --------------------------------------------------
Rv2715|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00862|M.leprae_Br4923 FLDTPPEEFDRVLAVNLGGVVNCCRAFGQRLVERGTGGHIVNVSSMAAYA
MAV_2275|M.avium_104 FLDTPAEEFDRVLAVNLGGVVNGCRSFARRMVQRGTGGHLVNVSSMAAYA
Mflv_4667|M.gilvum_PYR-GCK --------------------------------------------------
TH_0447|M.thermoresistible__bu --------------------------------------------------
MMAR_5090|M.marinum_M --------------------------------------------------
MUL_4166|M.ulcerans_Agy99 --------------------------------------------------
Mvan_5882|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_1510|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_1587|M.smegmatis_MC2_155 --------------------------------------------------
Mb2734|M.bovis_AF2122/97 --------------------------------------------------
Rv2715|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00862|M.leprae_Br4923 PLQSLSAYCTSKAATYMFSDCLRAELDAVGVGLTTICPGVIDTNIIQSTR
MAV_2275|M.avium_104 PLQSLSAYCTSKAATFMFSDCLRAELDAAGVGLTTICPGLIDTNIINTTR
Mflv_4667|M.gilvum_PYR-GCK --------------------------------------------------
TH_0447|M.thermoresistible__bu --------------------------------------------------
MMAR_5090|M.marinum_M --------------------------------------------------
MUL_4166|M.ulcerans_Agy99 --------------------------------------------------
Mvan_5882|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_1510|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_1587|M.smegmatis_MC2_155 --------------------------------------------------
Mb2734|M.bovis_AF2122/97 --------------------------------------------------
Rv2715|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00862|M.leprae_Br4923 IDTPT-AKQERVRDRRGQLDMMFRLRRYGPDKVADTILSAIKKNKPIRPV
MAV_2275|M.avium_104 FDAPAGARAERVDDRRGQLGKMFALRHYGPDKVADAILSSVQKKKPIRPV
Mflv_4667|M.gilvum_PYR-GCK --------------------------------------------------
TH_0447|M.thermoresistible__bu --------------------------------------------------
MMAR_5090|M.marinum_M --------------------------------------------------
MUL_4166|M.ulcerans_Agy99 --------------------------------------------------
Mvan_5882|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_1510|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_1587|M.smegmatis_MC2_155 --------------------------------------------------
Mb2734|M.bovis_AF2122/97 ---------------------------
Rv2715|M.tuberculosis_H37Rv ---------------------------
MLBr_00862|M.leprae_Br4923 APEAYALYGISRVLPQVLRSTARIRVI
MAV_2275|M.avium_104 APEAYALYGLSRVMPQGLRNAARLRVI
Mflv_4667|M.gilvum_PYR-GCK ---------------------------
TH_0447|M.thermoresistible__bu ---------------------------
MMAR_5090|M.marinum_M ---------------------------
MUL_4166|M.ulcerans_Agy99 ---------------------------
Mvan_5882|M.vanbaalenii_PYR-1 ---------------------------
MAB_1510|M.abscessus_ATCC_1997 ---------------------------
MSMEG_1587|M.smegmatis_MC2_155 ---------------------------