For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PTDLLTYHGGEGEPLVLVHGLMGRGSTWSRQLPWLTRLGAVYTYDAPWHRGRDVDDPYPISTERFVADLG EAVAELGGPARLVGHSMGALHSWCLAAERPGLVSALVVEDMAPDFVGRTTGPWEPWLHALPVEFDSADQV FAEFGPLAGQYFLEAFDRTATGWRLHGHTSRWIQIAAEWGTRDYWAQWRAVRQPALLIEAGNSVTPPGQM LAMAEVDRSTKYLHVPEAGHLVHDEAQLAYRQAVEAFLAAERVR*
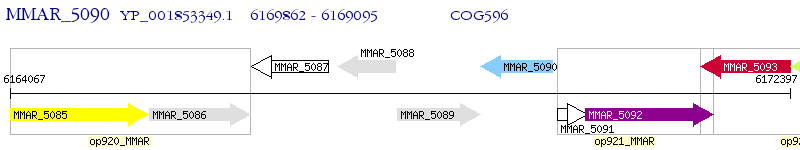
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5090 | - | - | 100% (255) | hydrolase |
| M. marinum M | MMAR_0900 | bpoC | 1e-08 | 27.34% (256) | bromoperoxidase BpoC |
| M. marinum M | MMAR_2050 | - | 9e-06 | 25.10% (263) | hypothetical protein MMAR_2050 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3622c | - | 1e-127 | 84.00% (250) | putative hydrolase |
| M. gilvum PYR-GCK | Mflv_1435 | - | 1e-122 | 80.48% (251) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv3591c | - | 1e-127 | 84.00% (250) | hydrolase |
| M. leprae Br4923 | MLBr_01921 | - | 1e-118 | 77.42% (248) | hypothetical protein MLBr_01921 |
| M. abscessus ATCC 19977 | MAB_0554 | - | 2e-95 | 64.43% (253) | alpha/beta family hydrolase |
| M. avium 104 | MAV_0564 | - | 1e-128 | 85.77% (246) | hydrolase, alpha/beta hydrolase fold family protein |
| M. smegmatis MC2 155 | MSMEG_6085 | - | 1e-111 | 73.71% (251) | hydrolase, alpha/beta hydrolase fold family protein |
| M. thermoresistible (build 8) | TH_0592 | - | 1e-111 | 75.81% (248) | POSSIBLE HYDROLASE |
| M. ulcerans Agy99 | MUL_4166 | - | 1e-153 | 99.61% (255) | hydrolase |
| M. vanbaalenii PYR-1 | Mvan_5351 | - | 1e-117 | 77.02% (248) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1435|M.gilvum_PYR-GCK ----MRTDLLTARGGA--GRPVVLVHGLMGRGTTWPRQLPWLIRHGRVYT
Mvan_5351|M.vanbaalenii_PYR-1 ------MDLLTFRGGE--GRPLVLVHGLMGRGSTWPRQLPWLTRLGRVFT
MMAR_5090|M.marinum_M ----MPTDLLTYHGGE--GEPLVLVHGLMGRGSTWSRQLPWLTRLGAVYT
MUL_4166|M.ulcerans_Agy99 ----MPTDLLTYHGGE--GEPLVLVHGLMGRGSTWSRQLPWLTRLGAVYT
MAV_0564|M.avium_104 --------MLTRHGGH--GEPLVLVHGLMGRGSTWTRQLPWLTRLGTVYS
Mb3622c|M.bovis_AF2122/97 -MPRMPANLLTHRGGR--GEPLVLVHGLMGRGSTWARQLPWLTLLGAVYT
Rv3591c|M.tuberculosis_H37Rv -MPRMPANLLTHRGGR--GEPLVLVHGLMGRGSTWARQLPWLTLLGAVYT
MLBr_01921|M.leprae_Br4923 ----MSTNLLTLRGGR--GKPLILVHGLMGRGNTWSRQLPWLSQLGTVYT
MSMEG_6085|M.smegmatis_MC2_155 ----MVTDLLTARGGH--GEPIVLVHGLMGRGTTWSRQLPWLTGLGEVYT
TH_0592|M.thermoresistible__bu MMSAVTGSLLTACGGASGGPPLVLVHGLMGRGSTWSRQVPWLARFGPVHT
MAB_0554|M.abscessus_ATCC_1997 -MSGMQRDHITDHGGHADGPTLVLVHGLMGRGTTWSRQLPWLRGLGRVYT
:* ** * .::*********.**.**:*** * *.:
Mflv_1435|M.gilvum_PYR-GCK YDAPWHRGREVDDPHPICTERFVADLGEAVESLGEPAILVGHSMGGLHSW
Mvan_5351|M.vanbaalenii_PYR-1 YDAPWHRGRDVADAHPIGTERFVADLGDAVQQLGEPALMVGHSMGALHSW
MMAR_5090|M.marinum_M YDAPWHRGRDVDDPYPISTERFVADLGEAVAELGGPARLVGHSMGALHSW
MUL_4166|M.ulcerans_Agy99 YDAPWHRGRDVDDPYPISTERFVADLGEAVAELGGPARLVGHSMGALHSW
MAV_0564|M.avium_104 YDAPWHRGRDVEDPHPISTERFVADLADAVGTLDAPARMVGHSMGALHSW
Mb3622c|M.bovis_AF2122/97 YDAPWHRGRDVADPHPISTERFVADLGDAVSALGAPTRMVGHSMGALHSW
Rv3591c|M.tuberculosis_H37Rv YDAPWHRGRDVADPHPISTERFVADLGDAVSALGAPTRMVGHSMGALHSW
MLBr_01921|M.leprae_Br4923 YDAPWHRGRNVVDPYPISTERFVTDLADAVSTLGVPARLVGHSMGAMHSW
MSMEG_6085|M.smegmatis_MC2_155 YDAPWHRGRGVADPHPISTERFVEDLTDAVGALGRPAILIGHSMGALHSW
TH_0592|M.thermoresistible__bu YDAPWHRGRDVLDPHPISTERFVDDLAAAVGELGGPAILIGHSMGGLHSW
MAB_0554|M.abscessus_ATCC_1997 YDAPWHRGGDVRDAQPVSTERYVDALAEAVRSVGAPVVLIGHSMGGLHSW
******** * *. *: ***:* * ** :. *. ::*****.:***
Mflv_1435|M.gilvum_PYR-GCK CLAAERPELVTALVVEDMAPDFRGRTTGPWEPWVHALPVEFTSADQVYAE
Mvan_5351|M.vanbaalenii_PYR-1 CLAAARPELVAGVVVEDMAPDFRGRTTGPWEPWVHALPVEFATADEVYAE
MMAR_5090|M.marinum_M CLAAERPGLVSALVVEDMAPDFVGRTTGPWEPWLHALPVEFDSADQVFAE
MUL_4166|M.ulcerans_Agy99 CLAAERPGLVSALVVEDMAPDFVGRTTGPWEPWLHALPVEFDSADQVFAE
MAV_0564|M.avium_104 CLAAQHPKLVSALVVEDMAPDFRGRTTGPWEPWLHALPVEFDSAEQVFAE
Mb3622c|M.bovis_AF2122/97 CLAAERPELVSALVVEDMAPDFRGRTTGPWEPWLRALPVEFDSAEQVFAE
Rv3591c|M.tuberculosis_H37Rv CLAAERPELVSALVVEDMAPDFRGRTTGPWEPWLRALPVEFDSAEQVFAE
MLBr_01921|M.leprae_Br4923 CLAAERPDLVSALVVEDMAPDFRGCSTGPWEPWLRGIPVEFDSVEKVFDE
MSMEG_6085|M.smegmatis_MC2_155 CLAAARPDLVTAVVVEDMAPDFRGRTTGPWEPWVHALPVEFASAQEVYDE
TH_0592|M.thermoresistible__bu CLAAARPDLVSAVVVEDMAPDFRGRTTGPWEPWLHALPVEFHSEQQVYDE
MAB_0554|M.abscessus_ATCC_1997 CLAAQRPDLVSALVVEDMAPDFVGGTTGPWEPWLYSWPEVFTSAGEFTAR
**** :* **:.:********* * :*******: . * * : :. .
Mflv_1435|M.gilvum_PYR-GCK FGPVAGQYFLEAFDRTATGWRLHGHPARWIEIAAEWGTRDYWSQWEAVTA
Mvan_5351|M.vanbaalenii_PYR-1 FGPVAGQYFLEAFDRTVTGWRLHGLPEKWIEIAAEWGTRDYWEQWRSVSV
MMAR_5090|M.marinum_M FGPLAGQYFLEAFDRTATGWRLHGHTSRWIQIAAEWGTRDYWAQWRAVRQ
MUL_4166|M.ulcerans_Agy99 FGPLAGQYFLEAFDHTATGWRLHGHTSRWIQIAAEWGTRDYWAQWRAVRQ
MAV_0564|M.avium_104 FGPVAGQYFLEAFDRTETGWRLHGHTSRWIEIAAEWGTRDYWAQWRAVRV
Mb3622c|M.bovis_AF2122/97 FGPVAGRYFLDAFDRTATGWRLHGRTARWIEIAAEWGTRDYWAQWRAVRS
Rv3591c|M.tuberculosis_H37Rv FGPVAGRYFLDAFDRTATGWRLHGRTARWIEIAAEWGTRDYWAQWRAVRS
MLBr_01921|M.leprae_Br4923 FGPVAGQYFLDAFDRTSTGWQLHGHTSRWIEIAAEWGTRDYWEHWLAVRS
MSMEG_6085|M.smegmatis_MC2_155 FGPVAGQYFLEAFDRTPTGWRLHGHPQWWIDIAAEWGRRDYWQQWRDVAA
TH_0592|M.thermoresistible__bu FGPIAGRYFLEAFDRTDTGWRLHGQPRFWIEIAAEWGVREYWEQWRSVRV
MAB_0554|M.abscessus_ATCC_1997 FGEIAGRYFREAFDRVDDGWRLHGPIEEWISTAAEWGVRDFWDQWRAVQC
** :**:** :***:. **:*** **. ***** *::* :* *
Mflv_1435|M.gilvum_PYR-GCK PTLLIEAGNSVTPPGQMRAMAQRSRGAEYLHVPGAGHLVHDDAPGKYRDA
Mvan_5351|M.vanbaalenii_PYR-1 PTLLLEAGASVTPPGQMRAMAEMAHRATYLHVPGAGHLIHDDAPGKYRDA
MMAR_5090|M.marinum_M PALLIEAGNSVTPPGQMLAMAEVDRSTKYLHVPEAGHLVHDEAQLAYRQA
MUL_4166|M.ulcerans_Agy99 PALLIEAGNSVTPPGQMLAMAEVDRSTKYLHVPEAGHLVHDEAQLAYRQA
MAV_0564|M.avium_104 PALLIEAGNSVTPPGQMLEMAERAVAATYLHVPQAGHLVHDEAPEAYRQA
Mb3622c|M.bovis_AF2122/97 PALLIEAGDGVTPPGQMRAMAERDYPTAYLRVPDAGHLVHDEAPQVYRRA
Rv3591c|M.tuberculosis_H37Rv PALLIEAGDGVTPPGQMRAMAERDYPTAYLRVPDAGHLVHDEAPQVYRRA
MLBr_01921|M.leprae_Br4923 PVLLIEAGTSVTPHGQMLAMTKTACRTTYLYVPEAGHLIHDEATQVYRQA
MSMEG_6085|M.smegmatis_MC2_155 PTLLLEAANSVAPAGQMRQMAETGAQTTYLYVSDAGHLIHDDQPEVYQAA
TH_0592|M.thermoresistible__bu PALLIEAGDSVTPPGQMRRMAECGHCARHVAVPGAGHLIHDDAPEAYRAA
MAB_0554|M.abscessus_ATCC_1997 PALLLEADDTVTPAGQMQKMSEIGRRTTYFKIAGSGHLMHDDSPKLYRSA
*.**:** *:* *** *:: : :. :. :***:**: *: *
Mflv_1435|M.gilvum_PYR-GCK VEAFLSSFA--------
Mvan_5351|M.vanbaalenii_PYR-1 VEAFLTSFTRDA-----
MMAR_5090|M.marinum_M VEAFLAAERVR------
MUL_4166|M.ulcerans_Agy99 VEAFLAAERVR------
MAV_0564|M.avium_104 VESFLAG----------
Mb3622c|M.bovis_AF2122/97 VESFLAGLTP-------
Rv3591c|M.tuberculosis_H37Rv VESFLAGLTP-------
MLBr_01921|M.leprae_Br4923 VVSFLAGCAGRD-----
MSMEG_6085|M.smegmatis_MC2_155 VTKFLAALAENAG----
TH_0592|M.thermoresistible__bu VEAFLAEFAECAGR---
MAB_0554|M.abscessus_ATCC_1997 VSDFLAAPGAPGASVVA
* **: