For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGTDHAELFTHLRPLLFTIAYEMLGSATEADDVLQESYLRWAQVDLDTVTDTKAYLAQIVSRQALNTLRT NSRRREEYVGPWLPEPVRVAYDDSDKDASTDVVLAESVSMAMLVVLETLAPDERAVFVMREVFGFDYSEI ASTVGKSEVAVRQMAHRARAHVQSRRRRFTPTDRQAEELTTKFLLAAAGGDVDALVEMLAPDAVWTSDGG GKVSAARRPVYGADRVARVAIGLSRAAAEFPDVRMEFSTFNSTPALLIYTEGRLDVVLAVSIEDGKVEHI FAIRNPDKLATAEITRHISR*
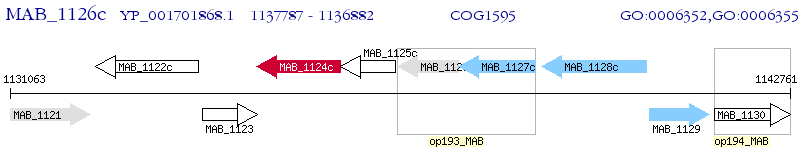
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1126c | - | - | 100% (301) | putative sigma factor |
| M. abscessus ATCC 19977 | MAB_4200 | - | 4e-95 | 57.62% (302) | putative sigma factor |
| M. abscessus ATCC 19977 | MAB_3678c | - | 7e-41 | 34.77% (302) | RNA polymerase sigma factor SigJ |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3361c | sigJ | 4e-45 | 36.11% (288) | RNA polymerase sigma factor SigJ |
| M. gilvum PYR-GCK | Mflv_1927 | - | 1e-104 | 64.67% (300) | ECF subfamily RNA polymerase sigma-24 factor |
| M. tuberculosis H37Rv | Rv3328c | sigJ | 4e-45 | 36.11% (288) | RNA polymerase sigma factor SigJ |
| M. leprae Br4923 | MLBr_01076 | sigE | 2e-07 | 23.95% (167) | RNA polymerase sigma factor SigE |
| M. marinum M | MMAR_4487 | - | 3e-98 | 61.74% (298) | alternative RNA polymerase sigma factor |
| M. avium 104 | MAV_1129 | - | 1e-99 | 60.98% (305) | sigma factor |
| M. smegmatis MC2 155 | MSMEG_5444 | - | 1e-105 | 64.78% (301) | RNA polymerase sigma-70 factor protein |
| M. thermoresistible (build 8) | TH_1142 | - | 1e-109 | 63.67% (300) | RNA polymerase sigma factor, sigma-70 family protein |
| M. ulcerans Agy99 | MUL_3630 | - | 8e-30 | 33.22% (304) | alternative RNA polymerase sigma factor SigJ- like |
| M. vanbaalenii PYR-1 | Mvan_4806 | - | 1e-105 | 64.88% (299) | ECF subfamily RNA polymerase sigma-24 factor |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3361c|M.bovis_AF2122/97 ------------MEVSEFEALRQHLMSVAYRLTGTVADAEDIVQEAWLRW
Rv3328c|M.tuberculosis_H37Rv ------------MEVSEFEALRQHLMSVAYRLTGTVADAEDIVQEAWLRW
Mflv_1927|M.gilvum_PYR-GCK -----MTAAPGDEHAERFTQLRPLLFTIAYEILGSATESDDVLQESYLRW
Mvan_4806|M.vanbaalenii_PYR-1 ----------MSEHADRFTHLRPLLFTIAYEILGSATESDDVLQESYLRW
MSMEG_5444|M.smegmatis_MC2_155 --MNNPSAGNSTEHAERFTALRPLLFTIAYEILGTATESDDVLQESYLRW
TH_1142|M.thermoresistible__bu MTGSGAPSARGDEHAERFTHLRPLLFTIAYEILGSATEADDVLQESYLRW
MMAR_4487|M.marinum_M ----MTPPSHSDEHAERFTLLRPLLFTIAYEILGSATESDDVLQESYLRW
MAV_1129|M.avium_104 ---MMTQAPTGSEHADRFTLLRPLLFTIAYEMLGSATEADDVLQDSYLRW
MAB_1126c|M.abscessus_ATCC_199 --------MSGTDHAELFTHLRPLLFTIAYEMLGSATEADDVLQESYLRW
MUL_3630|M.ulcerans_Agy99 --------MTDPALLARFQAARPRLGALAYRMLGSLADAEDVVQEAWLRL
MLBr_01076|M.leprae_Br4923 -----------------------MGDGIGMEREGRWTGN----TQCPLRV
:. . * : :. **
Mb3361c|M.bovis_AF2122/97 DSQD------TVIADPRAWLTTVVSRLGLD---KLRSAAHRRETYTGTWL
Rv3328c|M.tuberculosis_H37Rv DSPD------TVIADPRAWLTTVVSRLGLD---KLRSAAHRRETYTGTWL
Mflv_1927|M.gilvum_PYR-GCK AEVDL-----ATVTDTKAYLARLVTRQALN---TLRAQSRRREDYVGPWL
Mvan_4806|M.vanbaalenii_PYR-1 AEVDL-----AAVTDTKAYLARLVTRQALN---ALRAQSRRREDYVGPWL
MSMEG_5444|M.smegmatis_MC2_155 AEVDL-----ATVTDTKAYLAKLVTRQSLN---ALRARSRLREEYVGPWL
TH_1142|M.thermoresistible__bu AEVDL-----AAVRDTKSYLAKLVSRQALN---ALRAGARRREDYVGPWL
MMAR_4487|M.marinum_M AAVDL-----SAVRDTKSYLAQLVTRQSLN---ALRAGARRREEYVGPWL
MAV_1129|M.avium_104 STVDL-----ATVRDTKSYLAQLVTRQALN---ALRAGARRREEYVGPWL
MAB_1126c|M.abscessus_ATCC_199 AQVDL-----DTVTDTKAYLAQIVSRQALN---TLRTNSRRREEYVGPWL
MUL_3630|M.ulcerans_Agy99 NATTDRPEGAQELVNLDAWLTTVVARLCLN---LLRDRRGRGREELVGQL
MLBr_01076|M.leprae_Br4923 VPGDE-----SPTLDGRASPEDLIITNLLSPTIMSHPPPSRDDDWVEPFD
: : :: *. :
Mb3361c|M.bovis_AF2122/97 PEPVVTGLDAT--DPLAAVVAAEDARFAAMVVLERLRPDQRVAFVLHDGF
Rv3328c|M.tuberculosis_H37Rv PEPVVTGLDAT--DPLAAVVAAEDARFAAMVVLERLRPDQRVAFVLHDGF
Mflv_1927|M.gilvum_PYR-GCK PEPLLTEA-----DPSADVVLAESVSMAMLVVLETLSPDERAVFVLREVF
Mvan_4806|M.vanbaalenii_PYR-1 PEPLLTEA-----DPSADVVLAESVSMAMLVVLETLSPDERAVFVLREVF
MSMEG_5444|M.smegmatis_MC2_155 PEPLLVAE-----DASSDVVLAESVSMAMLVVLETLTPDERAVFVLHDVF
TH_1142|M.thermoresistible__bu PEPLLLEER----DASADVVLAESVSMAMMVVLESLTPDERAVFVLREVF
MMAR_4487|M.marinum_M PEPLLLDES----DPSADLVLAESVSMAMLVLLETLTPDERAVFVLREVF
MAV_1129|M.avium_104 PEPLLLDEQ----DPSTDVVLAESISMAMLVLLETLSPDERAVFVLREVF
MAB_1126c|M.abscessus_ATCC_199 PEPVRVAYDDSDKDASTDVVLAESVSMAMLVVLETLAPDERAVFVMREVF
MUL_3630|M.ulcerans_Agy99 PDPIVDGVAAF--DPEHHAMLADAVGLALFVVLDTLAPAERLAFVLHDVF
MLBr_01076|M.leprae_Br4923 -------------ALQGTAVFDATGDKATMPSWEELVRQH-----AARVY
: * : : * . :
Mb3361c|M.bovis_AF2122/97 AVPFAEVAEVLGTSEAAARQLASRARKAVTAQPALISG----DPDPAHNE
Rv3328c|M.tuberculosis_H37Rv AVPFAEVAEVLGTSEAAARQLASRARKAVTAQPALISG----DPDPAHNE
Mflv_1927|M.gilvum_PYR-GCK GFGHDEIAATLGKSTAAVRQIAHRAREHVQSRRKRFEP----VDPKTSGE
Mvan_4806|M.vanbaalenii_PYR-1 GFGHDEIAATLGKSTEAVRQTAHRARAHVQSRRKRFEP----VDPKVSTE
MSMEG_5444|M.smegmatis_MC2_155 GFSHDEIASTIGKSAAAVRQMAHRARNHVQARRKRFEP----VDPQVSME
TH_1142|M.thermoresistible__bu GFEYGEIATTVGKSVTAVRQIAHRARSHVHARRKRFEP----VDPEVSME
MMAR_4487|M.marinum_M GFDYDEIADAVGKPAPTVRQVAHRARAHIQARRKKYG----DVDPQRNAQ
MAV_1129|M.avium_104 GFDYDEIAEAVGKPAPTVRQVARRAREHVRARRKRHPGAGQAIDPKRNAE
MAB_1126c|M.abscessus_ATCC_199 GFDYSEIASTVGKSEVAVRQMAHRARAHVQSRRRRFTP-----TDRQAEE
MUL_3630|M.ulcerans_Agy99 AVPFDQIAPILERTPEATRKLASRARRRIERAEPFPDR-----DLCAQRE
MLBr_01076|M.leprae_Br4923 WLAYRLSGNQHDAEDLTQETFIRVFRSVQNYQPGTFEG--------WLHR
. . . : . * .
Mb3361c|M.bovis_AF2122/97 VVGRLMAAMAAGDLDTVVSLLHPDVTFTGDSNGKAPTAVRAVRGSDKVVR
Rv3328c|M.tuberculosis_H37Rv VVGRLMAAMAAGDLDTVVSLLHPDVTFTGDSNGKAPTAVRAVRGSDKVVR
Mflv_1927|M.gilvum_PYR-GCK ITMTFFAAAATGDLDALMAMLAPDVVWTADSDGKVSAARRPVEGADKVAR
Mvan_4806|M.vanbaalenii_PYR-1 ITTRFFTAAATGDLDGLMAMLAPDVVWTADSDGKVSAARRPVAGADRVAR
MSMEG_5444|M.smegmatis_MC2_155 ITQRFFTAASTGDIDALMELLAPDVVWTADSAGKRSAARRPVSGADAVAR
TH_1142|M.thermoresistible__bu ITERFLTAASTGDMDGLLAMLAPDATWTADGGGKATAARRPIVGAEKVAA
MMAR_4487|M.marinum_M LTAQFLQTAASGDVEALMAMLAPDATWLADSGGKVSAARKPVVGAERVAR
MAV_1129|M.avium_104 LTAQFLATAASGDVEALMAMLAPDATWTADSGGVVSAARRPVVGAEKVAR
MAB_1126c|M.abscessus_ATCC_199 LTTKFLLAAAGGDVDALVEMLAPDAVWTSDGGGKVSAARRPVYGADRVAR
MUL_3630|M.ulcerans_Agy99 VVDAFFAAGRSGNFERLVSVLHPDVVLRGDFG--PNTPRFRAQGAATVAK
MLBr_01076|M.leprae_Br4923 ITTNLFLDMVRRRARIRMDALPDDYDRVPADEPNPEQIYHDSRLGPDLQA
:. :: : * * . :
Mb3361c|M.bovis_AF2122/97 FILGLVQRYGPGLFGANQLALVNGELGAYTAGLPGVDGYRAMAPRITAIT
Rv3328c|M.tuberculosis_H37Rv FILGLVQRYGPGLFGANQLALVNGELGAYTAGLPGVDGYRAMAPRITAIT
Mflv_1927|M.gilvum_PYR-GCK LILGFVR-------LAG--GEARVEPAFYNSAPALVLYTGDVLEGVVTFE
Mvan_4806|M.vanbaalenii_PYR-1 LIMGFVR-------QAG--ADGRVEPAVYNSAPALVLYLGDNLEGVVTFE
MSMEG_5444|M.smegmatis_MC2_155 LVSGLVR-------LGG--EGARVEPAMYNNAPALKLYLGDSFEGVITIE
TH_1142|M.thermoresistible__bu VILGIFR-------AGQRLPDVRIETAIYNCAPAVVVYSRDNLEGVFTVE
MMAR_4487|M.marinum_M AITGLIR-------KAAT--ELQVELVTCNSAPAVLFYLGGQLEGVITLE
MAV_1129|M.avium_104 AITGLFR-------KAAEYATLRVDTVTCNGAPAVLLYLGDRLEGVITVE
MAB_1126c|M.abscessus_ATCC_199 VAIGLSR-------AAAEFPDVRMEFSTFNSTPALLIYTEGRLDVVLAVS
MUL_3630|M.ulcerans_Agy99 LARSYAG------------PERTVLNTLVNGAAGAVISISEQASAVMRFL
MLBr_01076|M.leprae_Br4923 ALDSLPP----------EFRAAVVLCDIEGLSYEEIGATLGVKLGTVRSR
.
Mb3361c|M.bovis_AF2122/97 VRDGK--VCALWDIANPDKFTGSPLKERRAQPTGRGRHHRN
Rv3328c|M.tuberculosis_H37Rv VRDGK--VCALWDIANPDKFTGSPLKERRAQPTGRGRHHRN
Mflv_1927|M.gilvum_PYR-GCK VADGR--ITNFYAMRNPEKLTGVMVARTISR----------
Mvan_4806|M.vanbaalenii_PYR-1 VVDGR--ITNFYAMRNPEKLAGVKVPRQISR----------
MSMEG_5444|M.smegmatis_MC2_155 IVDGR--ITHFYAMRNPDKLAGVDIPRVISR----------
TH_1142|M.thermoresistible__bu VVDGR--ITHFYAVRNPEKLAAVDTPRQISR----------
MMAR_4487|M.marinum_M IVGDK--ITNFYVMRNPDKLTAVARAHEISRG---------
MAV_1129|M.avium_104 IAADK--ITNFYVMRNPHKLAALATARDVSRG---------
MAB_1126c|M.abscessus_ATCC_199 IEDGK--VEHIFAIRNPDKLATAEITRHISR----------
MUL_3630|M.ulcerans_Agy99 VRNGR--IAAIDVLADPQRIATLDISGLSGPGG--------
MLBr_01076|M.leprae_Br4923 IHRGRQALRDYLLTAHPRDGATTDATHVKSA----------
: .: : .* : .