For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PGTPKTVVITGANAGIGLASAHRLAAGGHSIVMACRNQQKAQQARDQILAATPGAQIDIMSLDLSSFERI YRFAGELAAQHPQIDVLVNNAGASPMRQSKTDEGFELQWGANYLGPFLLTHLLLPQLQASRSGDARVIHL ASVAHSVGRINEKTWHGRRFYFTFSAYAQSKMGNLMFSNALARRLPDGVTSNALHPGNVRSEIWREIPQP VRSLVMLALITPERPSQLIADMAVSEEHRGRNGDYLAAQTPSPVRRYTKNLAAQDNLYAKSCALVGVEPL PVRG*
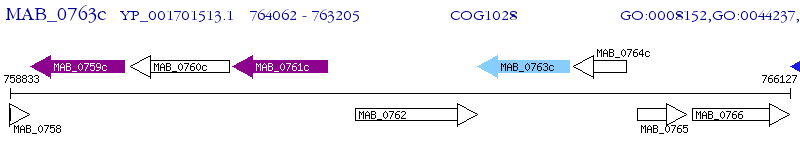
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0763c | - | - | 100% (285) | oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0646c | - | 1e-35 | 39.72% (214) | short chain dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0643 | - | 3e-35 | 34.95% (289) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0069 | - | 6e-40 | 35.76% (288) | short chain dehydrogenase |
| M. gilvum PYR-GCK | Mflv_0151 | - | 7e-33 | 40.38% (208) | short chain dehydrogenase |
| M. tuberculosis H37Rv | Rv0068 | - | 5e-40 | 35.76% (288) | short chain dehydrogenase |
| M. leprae Br4923 | MLBr_00315 | - | 1e-32 | 32.65% (291) | short chain dehydrogenase |
| M. marinum M | MMAR_3355 | - | 3e-36 | 41.38% (203) | oxidoreductase |
| M. avium 104 | MAV_4710 | - | 5e-36 | 41.10% (219) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_0863 | - | 5e-36 | 40.38% (208) | short chain dehydrogenase |
| M. thermoresistible (build 8) | TH_4179 | - | 1e-35 | 38.40% (250) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_1288 | - | 9e-36 | 40.89% (203) | short chain dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_0753 | - | 9e-37 | 36.77% (291) | short chain dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0151|M.gilvum_PYR-GCK ----MTS--KWTAADVPDQSGRVAVVTGANTGIGYETAEVLAGKGARVVI
Mvan_0753|M.vanbaalenii_PYR-1 ----MSS--KWTAADVPDQSGRVAVVTGANSGIGYEAAAVLAGRGARVVV
TH_4179|M.thermoresistible__bu ----MTG--KWTARDVPDQTGRTAVITGANTGLGFETAKVLAEKGAHVVL
MSMEG_0863|M.smegmatis_MC2_155 ----MSADTKWTEADVPDQSGRVAIVTGSNTGLGYETARALAAKGAHVVI
Mb0069|M.bovis_AF2122/97 -------MTKWTAADIPDQTGRTAVITGANTGLGFETAAALAAHGAHVVL
Rv0068|M.tuberculosis_H37Rv -------MTKWTAADIPDQTGRTAVITGANTGLGFETAAALAAHGAHVVL
MLBr_00315|M.leprae_Br4923 -------MAKWTTADIPDQTGRVAVITGANTGLGYQTALALAEHGAHVVL
MAV_4710|M.avium_104 MAPHDKRHRDWSEADVGDQSGRVVVITGANTGIGYETAAVLAHRGAHVVL
MMAR_3355|M.marinum_M -------MASDLIATVPDLSGKLAIVTGANSGLGFGLARRLSAAGADVVM
MUL_1288|M.ulcerans_Agy99 -------MASDLIATVPDLSGKLAIVTGANSGLGFGLARRLSAAGADVVM
MAB_0763c|M.abscessus_ATCC_199 ---------------MPG-TPKTVVITGANAGIGLASAHRLAAGGHSIVM
: . : : .::**:*:*:* * *: * :*:
Mflv_0151|M.gilvum_PYR-GCK AVRDAGKGQKALDAITRKHPGAAVSLQELDLSSLGSVRRATDALRSAHPR
Mvan_0753|M.vanbaalenii_PYR-1 AVRNLDKGRQAVSRIRQLHPGADVMLQELDLSSLASVRAAADDLRAAHPR
TH_4179|M.thermoresistible__bu AVRDPDKGRRAADRITAAAPHADVTVRQLDLTSLDNIRRAADDLRAGYPR
MSMEG_0863|M.smegmatis_MC2_155 AVRNLDKGRDAVDRIMASTPKADLKLQKLDVGSLDSVRTAADELKGAYPH
Mb0069|M.bovis_AF2122/97 AVRNLDKGKQAAARITEATPGAEVELQELDLTSLASVRAAAAQLKSDHQR
Rv0068|M.tuberculosis_H37Rv AVRNLDKGKQAAARITEATPGAEVELQELDLTSLASVRAAAAQLKSDHQR
MLBr_00315|M.leprae_Br4923 AVRNLDKGKDAAARITATSAQNNVALQELDLASLESVRAAAKQLRSDYDH
MAV_4710|M.avium_104 AVRDLEKGNAALSRIVAASPNADVTLQQLDLASLASVRSAAEALRAAYPR
MMAR_3355|M.marinum_M AIRNRAKGEAAIEEIRSAVPDAKLSIKALDLSSLASVAALGDQLNSEGRP
MUL_1288|M.ulcerans_Agy99 AIRNRAKGEAAIEEIRSAVPDAKLSTKALDLSSLASVAALGDQLNSEGRP
MAB_0763c|M.abscessus_ATCC_199 ACRNQQKAQQARDQILAATPGAQIDIMSLDLSSFERIYRFAGELAAQHPQ
* *: *.. * * . : **: *: : * .
Mflv_0151|M.gilvum_PYR-GCK IDLLINNAGVMYPP-KQVTRDGFELQFGTNHLGHFAFTGLLLDNLLDVP-
Mvan_0753|M.vanbaalenii_PYR-1 IDLLINNAGVMYPP-KQTTSDGFELQFGTNHLGHFALTGLLLDRLLPVE-
TH_4179|M.thermoresistible__bu IDLLINNAGVMYPP-RQTTRDGFELQFGTNHLGHFALTGQLLDNILPVD-
MSMEG_0863|M.smegmatis_MC2_155 IDLLINNAGVMYPP-KQTTVDGFELQFGTNHLGPFALTGLLIDHLLPVE-
Mb0069|M.bovis_AF2122/97 IDLLINNAGVMYTP-RQTTADGFEMQFGTNHLGHFALTGLLIDRLLPVA-
Rv0068|M.tuberculosis_H37Rv IDLLINNAGVMYTP-RQTTADGFEMQFGTNHLGHFALTGLLIDRLLPVA-
MLBr_00315|M.leprae_Br4923 IDLLINNAGVMWTP-KSTTKDGFELQFGTNHLGHFAFTGLLLDRLLPIV-
MAV_4710|M.avium_104 IDLLINNAGVMWTP-KQVTEDGFELQFGTNHLGHFALTGLLLDHLLGVR-
MMAR_3355|M.marinum_M IDILINNAGVMTPPERDTTADGFELQFGSNHLGHFALTAHVLPLLRAAQ-
MUL_1288|M.ulcerans_Agy99 IDILINNAGVMTPPERDTTADGFELQFGSNHLGHFALTAHVLPLLRAAQ-
MAB_0763c|M.abscessus_ATCC_199 IDVLVNNAGASPMR-QSKTDEGFELQWGANYLGPFLLTHLLLPQLQASRS
**:*:****. :. * :***:*:*:*:** * :* :: :
Mflv_0151|M.gilvum_PYR-GCK -GSRVVTVASLAHKNLADIHFDDLQWERKYNRVAAYGQSKLANLMFTYEL
Mvan_0753|M.vanbaalenii_PYR-1 -GSRVVSVASIAHNIQADIHFDDLQWERSYNRVAAYGQSKLANLMFTYTL
TH_4179|M.thermoresistible__bu -GSRVVTVASIAHRNMADIHFDDLQWERGYHRVAAYGQSKLANLMFAYEL
MSMEG_0863|M.smegmatis_MC2_155 -GSRVVAVASVAHRIRAKIHFEDLQWERRYNRVEAYGQSKLANLLFAYEL
Mb0069|M.bovis_AF2122/97 -GSRVVTISSVGHRIRAAIHFDDLQWERRYRRVAAYGQAKLANLLFTYEL
Rv0068|M.tuberculosis_H37Rv -GSRVVTISSVGHRIRAAIHFDDLQWERRYRRVAAYGQAKLANLLFTYEL
MLBr_00315|M.leprae_Br4923 -GSRVITVSSLSHRLFADIHFNDLQWECNYNRVAAYGQSKLANLLFTYEL
MAV_4710|M.avium_104 -DSRVVTVSSLGHRLRAAIHFDDLHWERRYDRVAAYGQSKLANLLFTYEL
MMAR_3355|M.marinum_M -GARVVSLSSLAAR-RGRIHFDDLQFEKSYAAMTAYGQSKLAVLMFAREL
MUL_1288|M.ulcerans_Agy99 -GARVVSLSSLAAR-RGRIHFDDLQFEKSYAAMTAYGQSKLAVLMFAREL
MAB_0763c|M.abscessus_ATCC_199 GDARVIHLASVAHS-VGRINEKTWHGRRFYFTFSAYAQSKMGNLMFSNAL
.:**: ::*:. . *: . : . * . **.*:*:. *:*: *
Mflv_0151|M.gilvum_PYR-GCK QRRLAARG-APTIAVAAHPGISNTELMRHVP--GTSLPGVMK--------
Mvan_0753|M.vanbaalenii_PYR-1 ARRLAAKG-APTIAVAAHPGISNTELMRHIP--GSQLPGFAW--------
TH_4179|M.thermoresistible__bu QRRLSAKN-APTISVAAHPGVSNTELTRYIP--GARLPGVSL--------
MSMEG_0863|M.smegmatis_MC2_155 QRRLAAAG-KPTISVAAHPGLSNTELMRHIP--GTGLPGYHQ--------
Mb0069|M.bovis_AF2122/97 QRRLAPG--GTTIAVASHPGVSNTELVRNMP--RPLVAVAAI--------
Rv0068|M.tuberculosis_H37Rv QRRLAPG--GTTIAVASHPGVSNTEVVRNMP--RPLVAVAAI--------
MLBr_00315|M.leprae_Br4923 QRRLATR--QTTIAVAAHPGGSRTELTRTLP--ALIAPIFSV--------
MAV_4710|M.avium_104 QRRLAAAPDAKTIAVAAHPGGSNTELARHLP--GIFRPVQAV--------
MMAR_3355|M.marinum_M DRRSRAAG-WGVMSNAAHPGLTKTNLQISGPSHGREKPALMQRFYTTSWR
MUL_1288|M.ulcerans_Agy99 DRRSRAAG-WGVMSNAAHPGLTKTNLQISGPSHGREKPALMQRFYTTSWR
MAB_0763c|M.abscessus_ATCC_199 ARRLPDG----VTSNALHPGNVRSEIWREIP-----QPVRSL-------V
** . : * *** .::: * .
Mflv_0151|M.gilvum_PYR-GCK LAGLVTNTPAVGALPTVRAATDPGVTGGQYYGPSGFNEMVGH-PVLVTSN
Mvan_0753|M.vanbaalenii_PYR-1 LAGLVTNSPAVGSLATLRAATDPGVRGGQYYGPSGVRELVGH-PVLVQSN
TH_4179|M.thermoresistible__bu LAGLLTNSPAVGALATLRAATDPEVKGGQYYGPDGFQEIRGH-PVLVGSS
MSMEG_0863|M.smegmatis_MC2_155 IASLFSNSPLMGALATLRAATDPGVKGGQYYGPDGFREVRGH-PELVKSS
Mb0069|M.bovis_AF2122/97 LAP-LMQDAELGALPTLRAATDPAVRGGQYFGPDGFGEIRGY-PKVVASS
Rv0068|M.tuberculosis_H37Rv LAP-LMQDAELGALPTLRAATDPAVRGGQYFGPDGFGEIRGY-PKVVASS
MLBr_00315|M.leprae_Br4923 AELFLTQDAATGALPTLRAATDAAVLGGQYFGPDGFAEIRGH-PKVVASN
MAV_4710|M.avium_104 LGPVLFQSPAMGALPTLRAATDPAVQGAQYYGPDGFLEQRGR-PKLVESS
MMAR_3355|M.marinum_M FAPFLWQEIEDGILPALYAAVTPQAEGGAFYGPRGFYEAAGGGVRAAKVP
MUL_1288|M.ulcerans_Agy99 FAPFLWQEIEDGILPALYAAVTPQAEGGAFYGPRGFYEAAGGGVRAAKVP
MAB_0763c|M.abscessus_ATCC_199 MLALITPERPSQLIADMAVSEEHRGRNGDYLAAQTPSPVRRY--------
. :. : .: .. : ..
Mflv_0151|M.gilvum_PYR-GCK KKSHDVAVQQRLWTVSEELTGVKYGV---
Mvan_0753|M.vanbaalenii_PYR-1 RKSHDVDVQERLWTVSEELTGVSYDL---
TH_4179|M.thermoresistible__bu AKSRDEDIQRRLWTVSEELTGVTFPV---
MSMEG_0863|M.smegmatis_MC2_155 SQSRDPELQRRLWAVSEELTGVSYPV---
Mb0069|M.bovis_AF2122/97 AQSHDEQLQRRLWAVSEELTGVVYPVG--
Rv0068|M.tuberculosis_H37Rv AQSHDEQLQRRLWAVSEELTGVVYPVG--
MLBr_00315|M.leprae_Br4923 GKSHDVDRQLRLWAVSEELTGVVYPVG--
MAV_4710|M.avium_104 AQSHDEQLQRRLWAVSEELTGVHFPV---
MMAR_3355|M.marinum_M ARAGNDADCQRLWEVSERLTGVGYPTSS-
MUL_1288|M.ulcerans_Agy99 ARAGNDADCQRLWEVSERLTGVGYPTSS-
MAB_0763c|M.abscessus_ATCC_199 --TKNLAAQDNLYAKSCALVGVEPLPVRG
: : .*: * *.**