For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSTEESSLGRGRRRSSDIDVKALAAARELLVEQGWEATTMVAIADRASVGKPALYRRWPSRAHLVFEAVF GWTAPTPEMSAAAGVTDWVRQSCSYTAELFERPDVIAAAPALLGQMRTDPELGQALWGSFGATGAGLLSR VMRAQQPEIGQEEAQERANAIMFMIIGANMLARQLLPGEHAEAVIKHLPELLTEPGESPGSVRPSG
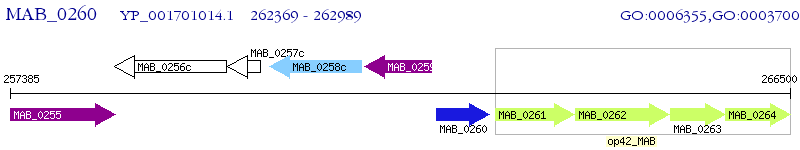
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0260 | - | - | 100% (206) | transcriptional regulator TetR |
| M. abscessus ATCC 19977 | MAB_0832 | - | 1e-09 | 31.33% (150) | TetR family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_3151 | - | 1e-08 | 47.37% (57) | TetR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3192c | - | 3e-20 | 35.67% (171) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0492 | - | 9e-20 | 36.14% (166) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3167c | - | 3e-20 | 35.67% (171) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_00815 | - | 8e-05 | 38.60% (57) | putative TetR-family transcriptional regulator |
| M. marinum M | MMAR_0340 | - | 1e-20 | 39.41% (170) | TetR family transcriptional regulator |
| M. avium 104 | MAV_5165 | - | 5e-21 | 37.84% (185) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_2043 | - | 2e-18 | 34.41% (186) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_0688 | - | 6e-17 | 34.10% (173) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY |
| M. ulcerans Agy99 | MUL_4780 | - | 9e-21 | 39.41% (170) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1870 | - | 3e-17 | 33.15% (178) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3192c|M.bovis_AF2122/97 -------------MKADLPSLDKAPGAGRPRDPRID-----SAILSATAE
Rv3167c|M.tuberculosis_H37Rv -------------MKADLPSLDKAPGAGRPRDPRID-----SAILSATAE
MSMEG_2043|M.smegmatis_MC2_155 -------------MKAEASPLGKGVGAGRPRDPRID-----AAILRATAE
Mvan_1870|M.vanbaalenii_PYR-1 -------------MNTDPSPGDKTPGAGRPRDPRID-----AAILRATAD
TH_0688|M.thermoresistible__bu -------------MTTNSTSVDKAAGAGRPRDPRID-----GAILRATAD
MMAR_0340|M.marinum_M -------------MVADF---------GRPRDPRID-----GAVLRATVE
MUL_4780|M.ulcerans_Agy99 -------------MVADF---------GRPRDPRID-----GAVLRATVE
MAV_5165|M.avium_104 ---------------MDF---------GRPRDPRID-----AAVLRATVE
Mflv_0492|M.gilvum_PYR-GCK -------------MSSDN---------GRPRDTRID-----SAVLDAAAE
MAB_0260|M.abscessus_ATCC_1997 -------------MSTEESSLGR----GRRRSSDID-----VKALAAARE
MLBr_00815|M.leprae_Br4923 MNNLAKAAQRRTVRPIDRGRLSDDIATPNQRSNRLSRDKRRGQLLIVASD
: . *. :. * .: :
Mb3192c|M.bovis_AF2122/97 LLVQIGYSNLSLAAVAERAGTTKSALYRRWSSKAELVHEAAFP---AAPT
Rv3167c|M.tuberculosis_H37Rv LLVQIGYSNLSLAAVAERAGTTKSALYRRWSSKAELVHEAAFP---AAPT
MSMEG_2043|M.smegmatis_MC2_155 LLVEIGYANLTMAAVAERAQTTKTALYRRWSSKAELVHEAVFP---AVPT
Mvan_1870|M.vanbaalenii_PYR-1 LLVEIGYSNVTMAAVAERAGTTKTALYRRWSSKAELVHEAAFP---VTPT
TH_0688|M.thermoresistible__bu LLVEIGYSNLTMSAVAERAGTTKTALYRRWSSKPELVYAATAP---TDPV
MMAR_0340|M.marinum_M LLAETGYAGLLVSAIAKRAGTSKPAIYRRWPSKAHLVHEAVFP---IDAA
MUL_4780|M.ulcerans_Agy99 LLAETGYAGLLVSAIAKRAGTSKPAIYRRWPSKAHLVHEAVFP---IDAA
MAV_5165|M.avium_104 LLAETGYPGLLVSAIAQRAGTSKPAIYRRWPSKAHLVHEAVFP---VGAD
Mflv_0492|M.gilvum_PYR-GCK LLLAVGYADLTVAAIAERANTSRPAVYRRWPSKAHLVHEAVFR---EADT
MAB_0260|M.abscessus_ATCC_1997 LLVEQGWEATTMVAIADRASVGKPALYRRWPSRAHLVFEAVFGW--TAPT
MLBr_00815|M.leprae_Br4923 VFVDHGYHAAGMDEIADRAGVSKPVLYQHFSSKLELYLAVLERHMNNLVS
:: *: : :*.** . :..:*:::.*: .* .
Mb3192c|M.bovis_AF2122/97 ALQAAAGDIAADIRMMIAATRDVFTTPVVRAALPGLVADMTADAELNARV
Rv3167c|M.tuberculosis_H37Rv ALQAAAGDIAADIRMMIAATRDVFTTPVVRAALPGLVADMTADAELNARV
MSMEG_2043|M.smegmatis_MC2_155 GLVAPAGDIAADVHAMIAAARDVFTSPVVRAALPGLIADVSADANLNARV
Mvan_1870|M.vanbaalenii_PYR-1 AIQTPPGDIAADLRAMIEAARDVFTSPVVRSALPGLIADLSADADLAGRV
TH_0688|M.thermoresistible__bu DTVESSDDVAGIVRALIERNRQITATPAARAAMPGLLVDMAADPRLHRGL
MMAR_0340|M.marinum_M TAVPDTGSPVDDLREMVRRTTAVLSTPAAKAALPGLIGEMAADPTLHSAL
MUL_4780|M.ulcerans_Agy99 TAVPDTGSPVDDLREMVRRTTAVLSTPAAKAAQPGLIGEMAADPTLHSAL
MAV_5165|M.avium_104 TAIPDTGSTADDLREMVRRTMVFLTTPAAKAALPGLIGEMAADPSLHSAL
Mflv_0492|M.gilvum_PYR-GCK SRVPVTGDLGRDLHSIVARTAALLTSPLARLAVPGLIGEAASDPSLNQRV
MAB_0260|M.abscessus_ATCC_1997 PEMSAAAGVTDWVRQSCSYTAELFERPDVIAAAPALLGQMRTDPELGQAL
MLBr_00815|M.leprae_Br4923 SVQQALRTTTNNRQRLHAAVQAFFDFIEHDGQGYRLIFENDNITEPRVAA
: . *: : .
Mb3192c|M.bovis_AF2122/97 LARFADLF-AAVRMRLREAVDR--GEAHPDVDPDRLIELIGGATMLRMLL
Rv3167c|M.tuberculosis_H37Rv LARFADLF-AAVRMRLREAVDR--GEAHPDVDPDRLIELIGGATMLRMLL
MSMEG_2043|M.smegmatis_MC2_155 MERFTGVF-AAVRERIVDGIAR--GEVHAAVSPDRLVEIIGGATMLRLLL
Mvan_1870|M.vanbaalenii_PYR-1 GARFTDLF-AAVRNRLHDAVER--GEVRAGVDPDQLVNLIGGATLLALLQ
TH_0688|M.thermoresistible__bu LGRVGSLV-EALRRRLAEAAGR--GEVQSGVDADRLTELIGGSVMFRMVL
MMAR_0340|M.marinum_M LVRFGEIFGHGLTTWLDNAAAR--GQVRADVTAPELAEAIAGIVLIGLLT
MUL_4780|M.ulcerans_Agy99 LVRFGEIFGHGLTTWLDNAAAR--GQVRADVTAPELAEAIAGIVLIGLLT
MAV_5165|M.avium_104 LERFAGAIGGGLADWLAAAAAR--GEARPDVTAAELAETIAGVTLVALLT
Mflv_0492|M.gilvum_PYR-GCK LDRFAGSGWSGLDAYLADAAAR--GELRAGFDAAALLEVVIGSALAAVLV
MAB_0260|M.abscessus_ATCC_1997 WGSFGATG-AGLLSRVMRAQQPEIGQEEAQERANAIMFMIIGANMLARQL
MLBr_00815|M.leprae_Br4923 QVRVATESCIDAVFALISADSR-LDPHRARMVAVGLVGISVDCARYWLDT
. : . . .. . : .
Mb3192c|M.bovis_AF2122/97 YPDDMLD----DAWVDQTTAIVVRGVHRAAPGGSVV
Rv3167c|M.tuberculosis_H37Rv YPDDMLD----DAWVDQTTAIVVRGVHRAAPGGSVV
MSMEG_2043|M.smegmatis_MC2_155 VSDENLD----DEWVHQTAAIVIHGVTVGR------
Mvan_1870|M.vanbaalenii_PYR-1 APDDLTA----PAWVDRTTDILLHGVMA--------
TH_0688|M.thermoresistible__bu HPDEPIT----ERWADEVAAILIRGVTKEG------
MMAR_0340|M.marinum_M R-PGELEPGVGDAWIDRTTTLLLKGIRKGTPQ----
MUL_4780|M.ulcerans_Agy99 R-PGELEPGVGDAWIDRTTTLLLKGIRKGTPQ----
MAV_5165|M.avium_104 R-PTELD----DAWVDRTTRLLLKGISA--------
Mflv_0492|M.gilvum_PYR-GCK RGPDGLT----DTWVSDTTAILLDGVRAR-------
MAB_0260|M.abscessus_ATCC_1997 LPGEHAE-----AVIKHLPELLTEPGESPGSVRPSG
MLBr_00815|M.leprae_Br4923 NRPISKS-----DAVEGTVQFAWGGLSHVPLTRL--
: