For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
FERFTDRARRVVVLAQEEARMLNHNYIGTEHILLGLIHEGEGVAAKSLESLGISLEGVRSQVEEIIGQGQ QAPSGHIPFTPRAKKVLELSLREALQLGHNYIGTEHILLGLIREGEGVAAQVLVKLGAELTRVRQQVIQL LSGYQGKEAAEAGTGGRGGESGSPSTSLVLDQFGRNLTAAAMEGKLDPVIGREKEIERVMQVLSRRTKNN PVLIGEPGVGKTAVVEGLAQAIVHGEVPETLKDKQLYTLDLGSLVAGSRYRGDFEERLKKVLKEINTRGD IILFIDELHTLVGAGAAEGAIDAASILKPKLARGELQTIGATTLDEYRKYIEKDAALERRFQPVQVGEPT VEHTIEILKGLRDRYEAHHRVSITDAAMVAAATLADRYINDRFLPDKAIDLIDEAGARMRIRRMTAPPDL REFDEKIAEARREKESAIDAQDFEKAASLRDREKTLVAQRAEREKQWRSGDLDVVAEVDDEQIAEVLGNW TGIPVFKLTEAETTRLLRMEEELHKRIIGQEDAVKAVSKAIRRTRAGLKDPKRPSGSFIFAGPSGVGKTE LSKALANFLFGDDDALIQIDMGEFHDRFTASRLFGAPPGYVGYEEGGQLTEKVRRKPFSVVLFDEIEKAH QEIYNSLLQVLEDGRLTDGQGRTVDFKNTVLIFTSNLGTSDISKPVGLGFSKGGGENDYERMKQKVNDEL KKHFRPEFLNRIDDIIVFHQLTREEIIRMVDLMISRVAGQLKSKDMALVLTDAAKALLAKRGFDPVLGAR PLRRTIQREIEDQLSEKILFEEVGPGQVVTVDVDNWDGEGPGEDAVFTFTGTRKPPAEPDLAKAGAHSAG GPEPAAR*
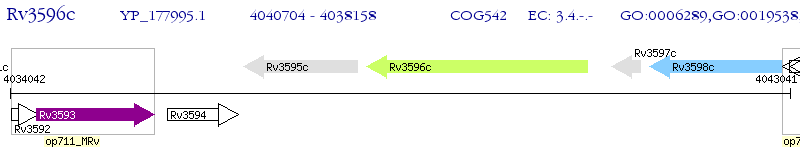
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv3596c | clpC1 | - | 100% (848) | PROBABLE ATP-DEPENDENT PROTEASE ATP-BINDING SUBUNIT CLPC1 |
| M. tuberculosis H37Rv | Rv0384c | clpB | 0.0 | 47.46% (866) | endopeptidase ATP binding protein |
| M. tuberculosis H37Rv | Rv2667 | clpC2 | 4e-20 | 38.61% (158) | ATP-dependent protease ATP-binding subunit ClpC2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3627c | clpC | 0.0 | 100.00% (848) | ATP-dependent Clp protease ATP-binding subunit CLPC |
| M. gilvum PYR-GCK | Mflv_1430 | - | 0.0 | 94.70% (849) | ATPase |
| M. leprae Br4923 | MLBr_00235 | clpC | 0.0 | 97.16% (845) | putative ATP-dependent Clp protease |
| M. abscessus ATCC 19977 | MAB_0546 | - | 0.0 | 92.56% (847) | ATP-dependent Clp protease ATP-binding subunit |
| M. marinum M | MMAR_5100 | clpC1 | 0.0 | 98.11% (845) | ATP-dependent protease ATP-binding subunit ClpC1 |
| M. avium 104 | MAV_0556 | - | 0.0 | 98.17% (819) | negative regulator of genetic competence ClpC/mecB |
| M. smegmatis MC2 155 | MSMEG_6091 | - | 0.0 | 95.38% (845) | negative regulator of genetic competence ClpC/mecB |
| M. thermoresistible (build 8) | TH_2346 | clpC | 0.0 | 93.97% (829) | PROBABLE ATP-DEPENDENT PROTEASE ATP-BINDING SUBUNIT CLPC1 |
| M. ulcerans Agy99 | MUL_4178 | clpC1 | 0.0 | 97.75% (846) | ATP-dependent protease ATP-binding subunit ClpC1 |
| M. vanbaalenii PYR-1 | Mvan_5358 | - | 0.0 | 94.81% (848) | ATPase |
CLUSTAL 2.0.9 multiple sequence alignment
Rv3596c|M.tuberculosis_H37Rv MFERFTDRARRVVVLAQEEARMLNHNYIGTEHILLGLIHEGEGVAAKSLE
Mb3627c|M.bovis_AF2122/97 MFERFTDRARRVVVLAQEEARMLNHNYIGTEHILLGLIHEGEGVAAKSLE
MLBr_00235|M.leprae_Br4923 MFERFTDRARRVVVLAQEEARMLNHNYIGTEHILLGLIHEGEGVAAKSLD
MMAR_5100|M.marinum_M MFERFTDRARRVVVLAQEEARMLNHNYIGTEHILLGLIHEGEGVAAKSLE
MUL_4178|M.ulcerans_Agy99 MFERFTDRARRVVVLAQEEARMLNHNYIGTEHILLGLIHEGEGVAAKSLE
MAV_0556|M.avium_104 ---------------------MLNHNYIGTEHILLGLIHEGEGVAAKSLE
Mflv_1430|M.gilvum_PYR-GCK MFERFTDRARRVVVLAQEEARMLNHNYIGTEHILLGLIHEGEGVAAKSLE
Mvan_5358|M.vanbaalenii_PYR-1 MFERFTDRARRVVVLAQEEARMLNHNYIGTEHILLGLIHEGEGVAAKSLE
MAB_0546|M.abscessus_ATCC_1997 MFERFTDRARRVVVLAQEEARMLNHNYIGTEHILLGLIHEGEGVAAKSLE
MSMEG_6091|M.smegmatis_MC2_155 MFERFTDRARRVVVLAQEEARMLNHNYIGTEHILLGLIHEGEGVAAKSLE
TH_2346|M.thermoresistible__bu MFERFTDRARRVVVLAQEEARMLNHNYIGTEHILLGLIHEGEGVAAKSLE
****************************:
Rv3596c|M.tuberculosis_H37Rv SLGISLEGVRSQVEEIIGQGQQAPSGHIPFTPRAKKVLELSLREALQLGH
Mb3627c|M.bovis_AF2122/97 SLGISLEGVRSQVEEIIGQGQQAPSGHIPFTPRAKKVLELSLREALQLGH
MLBr_00235|M.leprae_Br4923 SLGISLEAVRSQVEDIIGQGQQAPSGHIPFTPRAKKVLELSLREALQLGH
MMAR_5100|M.marinum_M SLGISLEGVRSQVEEIIGQGQQAPSGHIPFTPRAKKVLELSLREALQLGH
MUL_4178|M.ulcerans_Agy99 SLGISLEGVRSQVEEIIGQGQQAPSGHIPFTPRAKKVLELSLREALQLGH
MAV_0556|M.avium_104 SLGISLEGVRSQVEEIIGQGQQAPSGHIPFTPRAKKVLELSLREALQLGH
Mflv_1430|M.gilvum_PYR-GCK SLGISLEGVRSQVEEIIGQGQQAPSGHIPFTPRAKKVLELSLREALQLGH
Mvan_5358|M.vanbaalenii_PYR-1 SLGISLEGVRSQVEEIIGQGQQAPSGHIPFTPRAKKVLELSLREALQLGH
MAB_0546|M.abscessus_ATCC_1997 SLGISLEGVRSQVEEIIGQGQQAPSGHIPFTPRAKKVLELSLREALQLGH
MSMEG_6091|M.smegmatis_MC2_155 SLGISLEGVRSQVEEIIGQGQQAPSGHIPFTPRAKKVLELSLREALQLGH
TH_2346|M.thermoresistible__bu SLGISLEGVRSQVEEIIGQGQQAPSGHIPFTPRAKKVLELSLREALQLGH
*******.******:***********************************
Rv3596c|M.tuberculosis_H37Rv NYIGTEHILLGLIREGEGVAAQVLVKLGAELTRVRQQVIQLLSGYQGKEA
Mb3627c|M.bovis_AF2122/97 NYIGTEHILLGLIREGEGVAAQVLVKLGAELTRVRQQVIQLLSGYQGKEA
MLBr_00235|M.leprae_Br4923 NYIGTEHILLGLIREGEGVAAQVLVKLGAELTRVRQQVIQLLSGYQGKEA
MMAR_5100|M.marinum_M NYIGTEHILLGLIREGEGVAAQVLVKLGAELTRVRQQVIQLLSGYQGKEA
MUL_4178|M.ulcerans_Agy99 NYIGTEHILLGLIREGEGVAAQVLVKLGAELTRVRQQVIQLLSGYQGKEA
MAV_0556|M.avium_104 NYIGTEHILLGLIREGEGVAAQVLVKLGAELTRVRQQVIQLLSGYQGKEA
Mflv_1430|M.gilvum_PYR-GCK NYIGTEHILLGLIREGEGVAAQVLVKLGAELTRVRQQVIQLLSGYQGRET
Mvan_5358|M.vanbaalenii_PYR-1 NYIGTEHILLGLIREGEGVAAQVLVKLGAELTRVRQQVIQLLSGYQGKET
MAB_0546|M.abscessus_ATCC_1997 NYIGTEHILLGLIREGEGVAAQVLVKLGAELTRVRQQVIQLLSGYQGKET
MSMEG_6091|M.smegmatis_MC2_155 NYIGTEHILLGLIREGEGVAAQVLVKLGAELTRVRQQVIQLLSGYQGKEA
TH_2346|M.thermoresistible__bu NYIGTEHILLGLIREGEGVAAQVLVKLGAELTRVRQQVIQLLSGYQGKEA
***********************************************:*:
Rv3596c|M.tuberculosis_H37Rv AEAGTGGRGGESGSPSTSLVLDQFGRNLTAAAMEGKLDPVIGREKEIERV
Mb3627c|M.bovis_AF2122/97 AEAGTGGRGGESGSPSTSLVLDQFGRNLTAAAMEGKLDPVIGREKEIERV
MLBr_00235|M.leprae_Br4923 AEAGTGGRGGESGSPSTSLVLDQFGRNLTAAAMESKLDPVIGREKEIERV
MMAR_5100|M.marinum_M AEAGTGGRGGESGSPSTSLVLDQFGRNLTAAAMEGKLDPVIGREKEIERV
MUL_4178|M.ulcerans_Agy99 AEAGTGGRGGESGSPSTSLVLDQFGRNLTAAAMEGKLDPVIGREKEIERV
MAV_0556|M.avium_104 AEAGTGGRGGESGSPSTSLVLDQFGRNLTAAAMEGKLDPVIGREKEIERV
Mflv_1430|M.gilvum_PYR-GCK AEAGTGGRGGEAGNPSTSLVLDQFGRNLTAAAMEGKLDPVIGREKEIERV
Mvan_5358|M.vanbaalenii_PYR-1 AEAGTGGRGGEAGNPSTSLVLDQFGRNLTAAAMEGKLDPVIGREKEIERV
MAB_0546|M.abscessus_ATCC_1997 AESGTGGRGGEAGTPSTSLVLDQFGRNLTAAAMEGKLDPVIGREKEIERV
MSMEG_6091|M.smegmatis_MC2_155 AEAGTGGRGGESGNPSTSLVLDQFGRNLTAAAMEGKLDPVIGREKEIERV
TH_2346|M.thermoresistible__bu AEAGTGGRGGESGNPSTSLVLDQFGRNLTQAAAEGKLDPVIGREKEVERI
**:********:*.*************** ** *.***********:**:
Rv3596c|M.tuberculosis_H37Rv MQVLSRRTKNNPVLIGEPGVGKTAVVEGLAQAIVHGEVPETLKDKQLYTL
Mb3627c|M.bovis_AF2122/97 MQVLSRRTKNNPVLIGEPGVGKTAVVEGLAQAIVHGEVPETLKDKQLYTL
MLBr_00235|M.leprae_Br4923 MQVLSRRTKNNPVLIGEPGVGKTAVVEGLAQAIVHGEVPETLKDKQLYTL
MMAR_5100|M.marinum_M MQVLSRRTKNNPVLIGEPGVGKTAVVEGLAQAIVHGEVPETLKDKQLYTL
MUL_4178|M.ulcerans_Agy99 MQVLSRRTKNNPVLIGEPGVGKTAVVEGLAQAIVHGEVPETLKDKQLYTL
MAV_0556|M.avium_104 MQVLSRRTKNNPVLIGEPGVGKTAVVEGLAQAIVHGQVPETLKDKQLYTL
Mflv_1430|M.gilvum_PYR-GCK MQVLSRRTKNNPVLIGEPGVGKTAVVEGLAQAIVHGDVPETLKDKQLYTL
Mvan_5358|M.vanbaalenii_PYR-1 MQVLSRRTKNNPVLIGEPGVGKTAVVEGLAQAIVHGEVPETLKDKQLYTL
MAB_0546|M.abscessus_ATCC_1997 MQVLSRRTKNNPVLIGEPGVGKTAVVEGLAQAIVHGEVPETLKNKQLYTL
MSMEG_6091|M.smegmatis_MC2_155 MQVLSRRTKNNPVLIGEPGVGKTAVVEGLAQAIVHGEVPETLKDKQLYTL
TH_2346|M.thermoresistible__bu MQVLSRRTKNNPVLIGEPGVGKTAVVEGLAQAIVAGEVPETLKDKQLYTL
********************************** *:******:******
Rv3596c|M.tuberculosis_H37Rv DLGSLVAGSRYRGDFEERLKKVLKEINTRGDIILFIDELHTLVGAGAAEG
Mb3627c|M.bovis_AF2122/97 DLGSLVAGSRYRGDFEERLKKVLKEINTRGDIILFIDELHTLVGAGAAEG
MLBr_00235|M.leprae_Br4923 DLGSLVAGSRYRGDFEERLKKVLKEINTRGDIILFIDELHTLVGAGAAEG
MMAR_5100|M.marinum_M DLGSLVAGSRYRGDFEERLKKVLKEINTRGDIILFIDELHTLVGAGAAEG
MUL_4178|M.ulcerans_Agy99 DLGSLVAGSRYRGDFEERLKKVLKEINTRGDIILFIDELHTLVGAGAAEG
MAV_0556|M.avium_104 DLGSLVAGSRYRGDFEERLKKVLKEINTRGDIILFIDELHTLVGAGAAEG
Mflv_1430|M.gilvum_PYR-GCK DLGSLVAGSRYRGDFEERLKKVLKEINTRGDIILFIDELHTLVGAGAAEG
Mvan_5358|M.vanbaalenii_PYR-1 DLGSLVAGSRYRGDFEERLKKVLKEINTRGDIILFIDELHTLVGAGAAEG
MAB_0546|M.abscessus_ATCC_1997 DLGSLVAGSRYRGDFEERLKKVLKEINTRGDIILFIDELHTLVGAGAAEG
MSMEG_6091|M.smegmatis_MC2_155 DLGSLVAGSRYRGDFEERLKKVLKEINTRGDIILFIDELHTLVGAGAAEG
TH_2346|M.thermoresistible__bu DLGSLVAGSRYRGDFEERLKKVLKEINTRGDIILFIDELHTLVGAGAAEG
**************************************************
Rv3596c|M.tuberculosis_H37Rv AIDAASILKPKLARGELQTIGATTLDEYRKYIEKDAALERRFQPVQVGEP
Mb3627c|M.bovis_AF2122/97 AIDAASILKPKLARGELQTIGATTLDEYRKYIEKDAALERRFQPVQVGEP
MLBr_00235|M.leprae_Br4923 AIDAASILKPKLARGELQTIGATTLDEYRKYIEKDAALERRFQPVQVGEP
MMAR_5100|M.marinum_M AIDAASILKPKLARGELQTIGATTLDEYRKYIEKDAALERRFQPVQVGEP
MUL_4178|M.ulcerans_Agy99 AIDAASILKPKLARGELQTIGATTLDEYRKYIEKDAALERRFQPVQVGEP
MAV_0556|M.avium_104 AIDAASILKPKLARGELQTIGATTLDEYRKYIEKDAALERRFQPVQVGEP
Mflv_1430|M.gilvum_PYR-GCK AIDAASILKPKLARGELQTIGATTLDEYRKYIEKDAALERRFQPVQVGEP
Mvan_5358|M.vanbaalenii_PYR-1 AIDAASILKPKLARGELQTIGATTLDEYRKYIEKDAALERRFQPVQVGEP
MAB_0546|M.abscessus_ATCC_1997 AIDAASILKPKLARGELQTIGATTLDEYRKYIEKDAALERRFQPVQVGEP
MSMEG_6091|M.smegmatis_MC2_155 AIDAASILKPKLARGELQTIGATTLDEYRKYIEKDAALERRFQPVQVGEP
TH_2346|M.thermoresistible__bu AIDAASILKPKLARGELQTIGATTLDEYRKYIEKDAALERRFQPVQVGEP
**************************************************
Rv3596c|M.tuberculosis_H37Rv TVEHTIEILKGLRDRYEAHHRVSITDAAMVAAATLADRYINDRFLPDKAI
Mb3627c|M.bovis_AF2122/97 TVEHTIEILKGLRDRYEAHHRVSITDAAMVAAATLADRYINDRFLPDKAI
MLBr_00235|M.leprae_Br4923 TVEHTIEILKGLRDRYEAHHRVSITDSAMVAAATLADRYINDRFLPDKAI
MMAR_5100|M.marinum_M TVEHTIEILKGLRDRYEAHHRVSITDSAMVAAATLADRYINDRFLPDKAI
MUL_4178|M.ulcerans_Agy99 TVEHTIEILKGLRDRYEAHHRVSITDSAMVAAATLADRYINDRFLPDKAI
MAV_0556|M.avium_104 TVEHTIEILKGLRDRYEAHHRVSITDSAMVAAATLADRYINDRYLPDKAI
Mflv_1430|M.gilvum_PYR-GCK TVAHTIEILKGLRDRYEAHHRVSITDGAIAAAATLADRYINDRFLPDKAI
Mvan_5358|M.vanbaalenii_PYR-1 TVEHTIEILKGLRDRYEAHHRVSITDSALVAAATLADRYINDRFLPDKAI
MAB_0546|M.abscessus_ATCC_1997 TVEHTIEILKGLRDRYEAHHRVSITDGALVAAATLADRYINDRFLPDKAI
MSMEG_6091|M.smegmatis_MC2_155 TVEHTIEILKGLRDRYEAHHRVSITDSAMVAAATLADRYINDRFLPDKAI
TH_2346|M.thermoresistible__bu TVEHTIEILKGLRDRYEAHHRVSITDSAMVAAATLADRYINDRYLPDKAI
** ***********************.*:.*************:******
Rv3596c|M.tuberculosis_H37Rv DLIDEAGARMRIRRMTAPPDLREFDEKIAEARREKESAIDAQDFEKAASL
Mb3627c|M.bovis_AF2122/97 DLIDEAGARMRIRRMTAPPDLREFDEKIAEARREKESAIDAQDFEKAASL
MLBr_00235|M.leprae_Br4923 DLIDEAGARMRIRRMTAPPDLREFDEKIAEARREKESAIDAQDFEKAASL
MMAR_5100|M.marinum_M DLIDEAGARMRIRRMTAPPDLREFDEKIADARREKESAIDAQDFEKAASL
MUL_4178|M.ulcerans_Agy99 DLIDEAGARMRIRRMTAPPDLREFDEKIADARREKESAIDAQDFEKAASL
MAV_0556|M.avium_104 DLIDEAGARMRIRRMTAPPDLREFDEKIAEARREKESAIDAQDFEKAASL
Mflv_1430|M.gilvum_PYR-GCK DLIDEAGARMRIRRMTAPPDLREFDEKIADARREKESAIDAQDFEKAASL
Mvan_5358|M.vanbaalenii_PYR-1 DLIDEAGARMRIRRMTAPPDLREFDEKIADARREKESAIDAQDFEKAASL
MAB_0546|M.abscessus_ATCC_1997 DLIDEAGARMRIRRMTAPPDLREFDEKIADARREKESAIDAQDFEKAARL
MSMEG_6091|M.smegmatis_MC2_155 DLIDEAGARMRIRRMTAPPDLREFDEKIADARREKESAIDAQDFEKAAAL
TH_2346|M.thermoresistible__bu DLIDEAGARMRIRRMTAPPDLREFDEKIAETRREKESAIDAQDFEKAAAL
*****************************::***************** *
Rv3596c|M.tuberculosis_H37Rv RDREKTLVAQRAEREKQWRSGDLDVVAEVDDEQIAEVLGNWTGIPVFKLT
Mb3627c|M.bovis_AF2122/97 RDREKTLVAQRAEREKQWRSGDLDVVAEVDDEQIAEVLGNWTGIPVFKLT
MLBr_00235|M.leprae_Br4923 RDREKQLVAQRAEREKQWRSGDLDVIAEVDDEQIAEVLGNWTGIPVFKLT
MMAR_5100|M.marinum_M RDREKQLVAQRAEREKQWRSGDLDVVAEVDDEQIAEVLGNWTGIPVFKLT
MUL_4178|M.ulcerans_Agy99 RDREKQLVAQRAEREKQWRSGDLDVVAEVDDEQIAEVLGNWTGIPVFKLT
MAV_0556|M.avium_104 RDREKQLVAQRAEREKQWRSGDLDVVAEVDDEQIAEVLGNWTGIPVFKLT
Mflv_1430|M.gilvum_PYR-GCK RDKEKQLVAQRAEREKQWRSGDLDVVAEVDDEQIAEVLGNWTGIPVFKLT
Mvan_5358|M.vanbaalenii_PYR-1 RDKEKQLVAQRAEREKQWRSGDLDVVAEVDDEQIAEVLGNWTGIPVFKLT
MAB_0546|M.abscessus_ATCC_1997 RDSEKQLVAQRADREKQWRSGDLDVVAEVDDEQIAEVLGNWTGIPVFKLT
MSMEG_6091|M.smegmatis_MC2_155 RDKEKQLVAQRAEREKQWRSGDLDVVAEVDDEQIAEVLGNWTGIPVFKLT
TH_2346|M.thermoresistible__bu RDQEKQLVAQRAEREKQWRAGDLDVVAEVDDEQIAEVLGNWTGIPVFKLT
** ** ******:******:*****:************************
Rv3596c|M.tuberculosis_H37Rv EAETTRLLRMEEELHKRIIGQEDAVKAVSKAIRRTRAGLKDPKRPSGSFI
Mb3627c|M.bovis_AF2122/97 EAETTRLLRMEEELHKRIIGQEDAVKAVSKAIRRTRAGLKDPKRPSGSFI
MLBr_00235|M.leprae_Br4923 EAETTRLLRMEEELHKRIIGQEDAVKAVSKAIRRTRAGLKDPKRPSGSFI
MMAR_5100|M.marinum_M EAETTRLLRMEDELHKRIIGQEDAVKAVSKAIRRTRAGLKDPKRPSGSFI
MUL_4178|M.ulcerans_Agy99 EAETTRLLRMEDELHKRIIGQEDAVKAVSKAIRRTRAGLKDPKRPSGSFI
MAV_0556|M.avium_104 EAETTRLLRMEDELHKRIIGQEDAVKAVSKAIRRTRAGLKDPKRPSGSFI
Mflv_1430|M.gilvum_PYR-GCK EEETTRLLRMEDELHKRIIGQEDAVKAVSKAIRRTRAGLKDPKRPSGSFI
Mvan_5358|M.vanbaalenii_PYR-1 EEETTRLLRMEDELHKRIIGQEDAVKAVSKAIRRTRAGLKDPKRPSGSFI
MAB_0546|M.abscessus_ATCC_1997 EEETTRLLRMEDELHKRIIGQEDAVKAVSKAIRRTRAGLKDPRRPSGSFI
MSMEG_6091|M.smegmatis_MC2_155 EEETTRLLRMEEELHKRIIGQEDAVKAVSKAIRRTRAGLKDPKRPSGSFI
TH_2346|M.thermoresistible__bu EAETTRLLRMEEELHKRIIGQEDAVRAVSKAIRRTRAGLKDPKRPSGSFI
* *********:*************:****************:*******
Rv3596c|M.tuberculosis_H37Rv FAGPSGVGKTELSKALANFLFGDDDALIQIDMGEFHDRFTASRLFGAPPG
Mb3627c|M.bovis_AF2122/97 FAGPSGVGKTELSKALANFLFGDDDALIQIDMGEFHDRFTASRLFGAPPG
MLBr_00235|M.leprae_Br4923 FAGPSGVGKTELSKALANFLFGDDDALIQIDMGEFHDRFTASRLFGAPPG
MMAR_5100|M.marinum_M FAGPSGVGKTELSKALANFLFGDDDALIQIDMGEFHDRFTASRLFGAPPG
MUL_4178|M.ulcerans_Agy99 FAGPSGVGKTELSKALANFLFGDDDALIQIDMGEFHDRFTASRLFGAPPG
MAV_0556|M.avium_104 FAGPSGVGKTELSKALANFLFGDDDALIQIDMGEFHDRFTASRLFGAPPG
Mflv_1430|M.gilvum_PYR-GCK FAGPSGVGKTELSKALANFLFGDDDALIQIDMGEFHDRFTASRLFGAPPG
Mvan_5358|M.vanbaalenii_PYR-1 FAGPSGVGKTELSKALANFLFGDDDALIQIDMGEFHDRFTASRLFGAPPG
MAB_0546|M.abscessus_ATCC_1997 FAGPSGVGKTELSKALANFLFGDDDALIQIDMGEFHDRFTASRLFGAPPG
MSMEG_6091|M.smegmatis_MC2_155 FAGPSGVGKTELSKALANFLFGDDDALIQIDMGEFHDRFTASRLFGAPPG
TH_2346|M.thermoresistible__bu FAGPSGVGKTELSKALAEFLFGDEDALIQIDMGEYHDRFTASRLFGAPPG
*****************:*****:**********:***************
Rv3596c|M.tuberculosis_H37Rv YVGYEEGGQLTEKVRRKPFSVVLFDEIEKAHQEIYNSLLQVLEDGRLTDG
Mb3627c|M.bovis_AF2122/97 YVGYEEGGQLTEKVRRKPFSVVLFDEIEKAHQEIYNSLLQVLEDGRLTDG
MLBr_00235|M.leprae_Br4923 YVGYEEGGQLTEKVRRKPFSVVLFDEIEKAHQEIYNSLLQVLEDGRLTDG
MMAR_5100|M.marinum_M YVGYEEGGQLTEKVRRKPFSVVLFDEIEKAHQEIYNSLLQVLEDGRLTDG
MUL_4178|M.ulcerans_Agy99 YVGYEEGGQLTEKVRRKPFSVALFDEIEKAHQEISNSLLQVLEDGRLTDG
MAV_0556|M.avium_104 YVGYEEGGQLTEKVRRKPFSVVLFDEIEKAHQEIYNSLLQVLEDGRLTDG
Mflv_1430|M.gilvum_PYR-GCK YVGYEEGGQLTEKVRRKPFSVVLFDEIEKAHQEIYNSLLQVLEDGRLTDG
Mvan_5358|M.vanbaalenii_PYR-1 YVGYEEGGQLTEKVRRKPFSVVLFDEIEKAHQEIYNSLLQVLEDGRLTDG
MAB_0546|M.abscessus_ATCC_1997 YVGYEEGGQLTEKVRRKPFSVVLFDEIEKAHQEIYNTLLQVLEDGRLTDG
MSMEG_6091|M.smegmatis_MC2_155 YVGYEEGGQLTEKVRRKPFSVVLFDEIEKAHQEIYNSLLQVLEDGRLTDG
TH_2346|M.thermoresistible__bu YVGYEEGGQLTEKVRRKPFSVVLFDEIEKAHQEIYNTLLQVLEDGRLTDG
*********************.************ *:*************
Rv3596c|M.tuberculosis_H37Rv QGRTVDFKNTVLIFTSNLGTSDISKPVGLGFSKGGGENDYERMKQKVNDE
Mb3627c|M.bovis_AF2122/97 QGRTVDFKNTVLIFTSNLGTSDISKPVGLGFSKGGGENDYERMKQKVNDE
MLBr_00235|M.leprae_Br4923 QGRTVDFKNTVLIFTSNLGTSDISKPVGLGFTQGSGENDYERMKQKVNDE
MMAR_5100|M.marinum_M QGRTVDFKNTVLIFTSNLGTSDISKPVGLGFSQTGGENDYERMKQKVNDE
MUL_4178|M.ulcerans_Agy99 QGRTVDFKNTVLIFTSNLGTSDISKPVGLGFSQTGGENDYERMKQKVNDE
MAV_0556|M.avium_104 QGRTVDFKNTVLIFTSNLGTSDISKPVGLGFTQGGGENNYERMKQKVNDE
Mflv_1430|M.gilvum_PYR-GCK QGRTVDFKNTVLIFTSNLGTSDISKAVGLGFTQGGGENNYERMKQKVHDE
Mvan_5358|M.vanbaalenii_PYR-1 QGRTVDFKNTVLIFTSNLGTSDISKAVGLGFTQGGGENNYERMKQKVHDE
MAB_0546|M.abscessus_ATCC_1997 QGRTVDFKNTVLIFTSNLGTSDISKAVGLGFSEGAAGSNYERMKQKVHDE
MSMEG_6091|M.smegmatis_MC2_155 QGRTVDFKNTVLIFTSNLGTSDISKAVGLGFSQGGSENNYERMKQKVHDE
TH_2346|M.thermoresistible__bu QGRTVDFKNTVLIFTSNLGTSDISKAVGLGFSQGGSENNYERMKQKVHDE
*************************.*****:: .. .:********:**
Rv3596c|M.tuberculosis_H37Rv LKKHFRPEFLNRIDDIIVFHQLTREEIIRMVDLMISRVAGQLKS-KDMAL
Mb3627c|M.bovis_AF2122/97 LKKHFRPEFLNRIDDIIVFHQLTREEIIRMVDLMISRVAGQLKS-KDMAL
MLBr_00235|M.leprae_Br4923 LKKHFRPEFLNRIDDIIVFHQLSRDEIIRMVDLMISRVANQLKV-KDMTL
MMAR_5100|M.marinum_M LKKHFRPEFLNRIDDIIVFHQLTRDEIIRMVDLMIGRVANQLKS-KDMAL
MUL_4178|M.ulcerans_Agy99 LKKHFRPEFLNRIDDIIVFHQLTRDEIIRMVDLMIGRVANQLKSSKDMAL
MAV_0556|M.avium_104 LKKHFRPEFLNRIDDIIVFHQLTRDEIIRMVDLMITRVAGQLKS-KDMDL
Mflv_1430|M.gilvum_PYR-GCK LKKHFRPEFLNRIDDIIVFHQLTQDEIIQMVDLMIGRVGNQLKA-KDMAM
Mvan_5358|M.vanbaalenii_PYR-1 LKKHFRPEFLNRIDDIIVFHQLTQDEIIKMVDLMIGRVSKQLAA-KDMTM
MAB_0546|M.abscessus_ATCC_1997 LKKHFRPEFLNRIDDIIVFEQLTQEQIIEMVDLMIGRVGKQLKT-KDMAM
MSMEG_6091|M.smegmatis_MC2_155 LKKHFRPEFLNRIDDIIVFHQLTQDEIIQMVDLMIGRVSNQLKT-KDMAL
TH_2346|M.thermoresistible__bu LKKHFRPEFLNRIDDIIVFHQLTQEQIIQMVDLMIDRVRNQLKA-KDMDL
*******************.**::::**.****** ** ** *** :
Rv3596c|M.tuberculosis_H37Rv VLTDAAKALLAKRGFDPVLGARPLRRTIQREIEDQLSEKILFEEVGPGQV
Mb3627c|M.bovis_AF2122/97 VLTDAAKALLAKRGFDPVLGARPLRRTIQREIEDQLSEKILFEEVGPGQV
MLBr_00235|M.leprae_Br4923 ELTNKAKALLAKRGFDPVLGARPLRRTIQREIEDQLSEKILFEEVGPGQV
MMAR_5100|M.marinum_M ELTDKAKSLLAKRGFDPVLGARPLRRTIQREIEDQLSEKILFEEVGPGQV
MUL_4178|M.ulcerans_Agy99 ELTDKAKSLLAKRGFDPVLGARPLRRTIQREIEDQLSEKILFEEVGPGQV
MAV_0556|M.avium_104 VLTDKAKALLAKRGFDPVLGARPLRRTIQREIEDQLSEKILFEEVGPGQI
Mflv_1430|M.gilvum_PYR-GCK ELTPKAKALLAKRGFDPVLGARPLRRTIQREIEDQLSEKILFEELGPGQL
Mvan_5358|M.vanbaalenii_PYR-1 ELTDKAKSLLAKRGFDPVLGARPLRRTIQREIEDQLSEKILFEEVGPGQL
MAB_0546|M.abscessus_ATCC_1997 ELTDKAKSLLAKRGFDPVLGARPLRRTIQREIEDALSEKILFNEVGPGEL
MSMEG_6091|M.smegmatis_MC2_155 ELSDKAKALLAKRGFDPVLGARPLRRTIQREIEDQLSEKILFEEIGPGQL
TH_2346|M.thermoresistible__bu QLTDKAKQLLAKRGFDPVLGARPLRRTIQREIEDQLSEKILFEELRPGQL
*: ** ************************** *******:*: **::
Rv3596c|M.tuberculosis_H37Rv VTVDVDNWDGEGPGEDAVF-------------------------------
Mb3627c|M.bovis_AF2122/97 VTVDVDNWDGEGPGEDAVF-------------------------------
MLBr_00235|M.leprae_Br4923 VTVDVDNWDGEGPGEDVKF-------------------------------
MMAR_5100|M.marinum_M VTVDVDNWDGEGAGEDAKF-------------------------------
MUL_4178|M.ulcerans_Agy99 VTVDVDNWDGEGAGEDAKF-------------------------------
MAV_0556|M.avium_104 VTVDVDNWDGESAGEDAVF-------------------------------
Mflv_1430|M.gilvum_PYR-GCK VTVDVENWDGEGQGEDAVF-------------------------------
Mvan_5358|M.vanbaalenii_PYR-1 VTVDVEGWDGEGAGEDAKF-------------------------------
MAB_0546|M.abscessus_ATCC_1997 VTVDVENWDGESAGEDAKF-------------------------------
MSMEG_6091|M.smegmatis_MC2_155 VTVDVEGWDGEGQGEDAKF-------------------------------
TH_2346|M.thermoresistible__bu ITVDVENWDGEGAGEDAXXXPTPPTGPAADVLNQATRVVPPAAPPSGTSA
:****:.****. ***.
Rv3596c|M.tuberculosis_H37Rv --------------------TFTGTRKPPAEPDLAKAGAHSAGGPEPAAR
Mb3627c|M.bovis_AF2122/97 --------------------TFTGTRKPPAEPDLAKAGAHSAGGPEPAAR
MLBr_00235|M.leprae_Br4923 --------------------TFTGIRKPSTEPDLAKAGVHSAGGPEPVEQ
MMAR_5100|M.marinum_M --------------------TFTGTRKPPSEPDLAKAGAHSAGGPGPTEQ
MUL_4178|M.ulcerans_Agy99 --------------------TFTGTRKPPSEPDLAKAGAHSAGGPGPTEQ
MAV_0556|M.avium_104 --------------------TFTGTRKPPAEPDLAKAGAHSADPQ-----
Mflv_1430|M.gilvum_PYR-GCK --------------------TFSGARKPVEVAEPDLAQAGAGGAGPAAE-
Mvan_5358|M.vanbaalenii_PYR-1 --------------------TFIGTKKPVKIEEPDLAQAGAAGGPSAAE-
MAB_0546|M.abscessus_ATCC_1997 --------------------TFVGRPKPALVAG---EEPAADLAASASE-
MSMEG_6091|M.smegmatis_MC2_155 --------------------TFSGGPKRAETAEPDLAGAGAAGAPTAGTE
TH_2346|M.thermoresistible__bu ARSGLLPSLLLPLQTTANTVTSVVPPPPPPAPGTPEAAVLETASGSASAA
*
Rv3596c|M.tuberculosis_H37Rv --------------------------------------------------
Mb3627c|M.bovis_AF2122/97 --------------------------------------------------
MLBr_00235|M.leprae_Br4923 --------------------------------------------------
MMAR_5100|M.marinum_M --------------------------------------------------
MUL_4178|M.ulcerans_Agy99 --------------------------------------------------
MAV_0556|M.avium_104 --------------------------------------------------
Mflv_1430|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5358|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_0546|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6091|M.smegmatis_MC2_155 --------------------------------------------------
TH_2346|M.thermoresistible__bu MPGVAAATSTLFSPPELNLPSIPGVGIPLPNRLSLPTDGICEGLGWSAGK
Rv3596c|M.tuberculosis_H37Rv -----------------------
Mb3627c|M.bovis_AF2122/97 -----------------------
MLBr_00235|M.leprae_Br4923 -----------------------
MMAR_5100|M.marinum_M -----------------------
MUL_4178|M.ulcerans_Agy99 -----------------------
MAV_0556|M.avium_104 -----------------------
Mflv_1430|M.gilvum_PYR-GCK -----------------------
Mvan_5358|M.vanbaalenii_PYR-1 -----------------------
MAB_0546|M.abscessus_ATCC_1997 -----------------------
MSMEG_6091|M.smegmatis_MC2_155 -----------------------
TH_2346|M.thermoresistible__bu SDGQGAEVPAAPVPADRRDRWDR