For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLHDRLTGAPRGATGDEGAANAHITRAMVAALTSVATQIKTLDAQIAEQLSLHADAHIFTSLPRSGTVRA ARLLAEIGDCRARFPTPESLACLAGVAPSTRQSGKVKHVGFRWAADKQLRDAVCDFAGDSRRANLWAADR YNRAIARGHDHPHAVRILARAWLYAIWHCWQDGAAYHPANHRALQALLNQDQDRAA
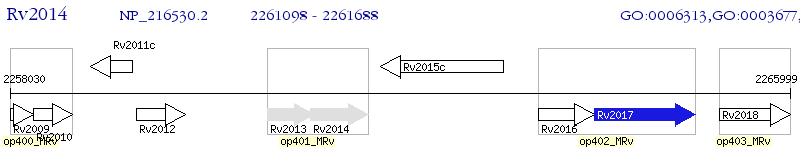
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2014 | - | - | 100% (196) | POSSIBLE TRANSPOSASE |
| M. tuberculosis H37Rv | Rv2177c | - | 9e-09 | 25.12% (203) | transposase |
| M. tuberculosis H37Rv | Rv2424c | - | 1e-08 | 26.42% (212) | transposase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2037 | - | 1e-113 | 100.00% (196) | transposase |
| M. gilvum PYR-GCK | Mflv_1197 | - | 1e-15 | 32.78% (180) | transposase, IS111A/IS1328/IS1533 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_3782 | - | 1e-14 | 32.48% (157) | transposase |
| M. avium 104 | MAV_0496 | - | 2e-14 | 35.10% (151) | transposase IS116/IS110/IS902 |
| M. smegmatis MC2 155 | MSMEG_1732 | - | 1e-14 | 31.76% (170) | transposase IS116/IS110/IS902 |
| M. thermoresistible (build 8) | TH_3366 | - | 8e-47 | 50.00% (184) | PUTATIVE transferase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4491 | - | 2e-14 | 31.21% (173) | transposase, IS111A/IS1328/IS1533 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv2014|M.tuberculosis_H37Rv --------------------------------------------------
Mb2037|M.bovis_AF2122/97 --------------------------------------------------
TH_3366|M.thermoresistible__bu ----------------------VGKILAQFMIDHTADGIATLIRKLSKFG
Mflv_1197|M.gilvum_PYR-GCK -MTLFIGDDWAEDHHDIHLMDAEGTRLASRRLPEGLAGIRGFHELVAAHA
Mvan_4491|M.vanbaalenii_PYR-1 --MIFVGDDWAEDHHDVYLMDESGQRLAARRLPEGLSGIRALHELIAGYA
MSMEG_1732|M.smegmatis_MC2_155 ---------------------------------------------MAAHC
MAV_0496|M.avium_104 -------------------------MVARGRVSNDAAGFAALLTLLAEAG
MMAR_3782|M.marinum_M MPTVWAGVDTGKRTHHCVVLDRNGGVLLSRPVDNDETALLELIEAVVGVA
Rv2014|M.tuberculosis_H37Rv --------------------------------------------------
Mb2037|M.bovis_AF2122/97 --------------------------------------------------
TH_3366|M.thermoresistible__bu DPADV--HIGIERPNGRLVDLLLEAGHPVIPVSPNAIKTWRDGEVISGAK
Mflv_1197|M.gilvum_PYR-GCK QDPTE-VVIGIETDRGLWVEALAAAGYEVFAVNPLAVARYRDRHQVSGAK
Mvan_4491|M.vanbaalenii_PYR-1 EEPDQ-VIVGIETDRGLWVSALAAAGYRVWAINPMAAARYRDRHHVSGAK
MSMEG_1732|M.smegmatis_MC2_155 EDSEQ-VVVGIETDNGLWVQALHAVGYRVYGINPLSASRYRDRHCVSGAK
MAV_0496|M.avium_104 DSAAHPIPVGIETDRGLWVAALRETGRVIYPINPLAASRYRARHAVSGAK
MMAR_3782|M.marinum_M AGREVCWATDLNAGGAAMLITLLAAHDQQLLYIPGRIVHHAAATYRGDGK
Rv2014|M.tuberculosis_H37Rv --------------------------------------------------
Mb2037|M.bovis_AF2122/97 --------------------------------------------------
TH_3366|M.thermoresistible__bu SDAGDALVIAEYLRLRHHRLRPAAPYSSSTKALRTVVRTRDDVVAMRVAA
Mflv_1197|M.gilvum_PYR-GCK SDAADAKLLADLVRTDRHNHRPIAGDTPEVEAIKVLARAHQNLIWSRNRN
Mvan_4491|M.vanbaalenii_PYR-1 SDASDAKLLADLVRTDRHNHREIAGDRPQAEAIKVLARSHQSLIWARTRH
MSMEG_1732|M.smegmatis_MC2_155 SDAGDAKILADLVRTDRHNHRQLADDSSAVHAIKVLARTHQSLVWARTRH
MAV_0496|M.avium_104 SDATDAVLLANIVRTDAAAHRPLPADTELAQAIRVLARAQQDAVWARQQI
MMAR_3782|M.marinum_M TDAKNARIIADQARMR-TDLQPVRRADPIATDLRLLTSRRTDVVCDRVRA
Rv2014|M.tuberculosis_H37Rv --------------------------------------------------
Mb2037|M.bovis_AF2122/97 --------------------------------------------------
TH_3366|M.thermoresistible__bu TNQLSALLDDHWPGAKAIFADVES-----PISLAFLRRYPTAASAARLGE
Mflv_1197|M.gilvum_PYR-GCK TNALRSALREYYPAALEAFASLSD-----RDALAILGRAPTPTDAARLSV
Mvan_4491|M.vanbaalenii_PYR-1 ANMLRSGLREYYPAALEAFDSLTD-----SDALAILGRAPTPDQGARLSL
MSMEG_1732|M.smegmatis_MC2_155 SNALRAGLSQYYPAALAAFPDIDG-----RDALAVLQRAPDPSAGATLSL
MAV_0496|M.avium_104 GNQIRDLLKDFYPAAIAAFAGLPEGGLARADARTILAAAPTPTQAATLTP
MMAR_3782|M.marinum_M INRLRQTLLEYFPPLERAFDYSKS-----KAALTLLSRYQTPDSLRRIGP
Rv2014|M.tuberculosis_H37Rv -------------------MLHDRLTGAPRG---ATG-DEGAANAHITRA
Mb2037|M.bovis_AF2122/97 -------------------MLHDRLTGAPRG---ATG-DEGAANAHITRA
TH_3366|M.thermoresistible__bu KRLAAFLVKHSYSGRRPVKELLTRLRSAPAG---TTDPVLTIAVRDVVLA
Mflv_1197|M.gilvum_PYR-GCK SKIRSALKAAG--RQRNLDAKALEIQTALRSDQLAAAPAVTAAFGATTRA
Mvan_4491|M.vanbaalenii_PYR-1 AKIGSALKSAG--RQRNIESRAAEIQTALRGEHLTAPPAVAAAFGATTAA
MSMEG_1732|M.smegmatis_MC2_155 SKISSALKVAG--RQRSIDARAREIQTALRSPQLHTSDQVVAALSASTTA
MAV_0496|M.avium_104 ARLRRLLTKAG--RRRHLDRDIERLRAVFTDTYLHQPPAVENAMGIQLAA
MMAR_3782|M.marinum_M TRLATWLKARG-----CRNSATVAQTAVQAAQTQHTTLPTQTVGAQLVPR
. .
Rv2014|M.tuberculosis_H37Rv MVAALTSVATQIKTLDAQIAEQLSLHADAHIFTSLPRSGTVRAARLLAEI
Mb2037|M.bovis_AF2122/97 MVAALTSVATQIKTLDAQIAEQLSLHADAHIFTSLPRSGTVRAARLLAEI
TH_3366|M.thermoresistible__bu LVSVIEALNSAGKALDRSVIARLGEHPDAEVFTSLPRSGQINAAQVLAEW
Mflv_1197|M.gilvum_PYR-GCK AVAIITELNHHIADLEAELAAHFETHPDADIYLSLPGLGVILGARVLGEF
Mvan_4491|M.vanbaalenii_PYR-1 AVHVITALNDQIAELETALADHFEAHPDADIYRSLPGLGVILGARVLGEF
MSMEG_1732|M.smegmatis_MC2_155 TVAVLLVLNTQISELQTQLTDHFETHPDADIYLSLPGLGVILGARVLGEF
MAV_0496|M.avium_104 LLRQLEAACTAADELAEAAIAHFEQHPDAKIITSFPGLGMLAGARVLAEI
MMAR_3782|M.marinum_M LAAEITTIDAELAHVDNEITARFADHDSAEILTSMPGFGPVLAATFLAQI
: : :: * .*.: *:* * : .* .*.:
Rv2014|M.tuberculosis_H37Rv GDCRARFPTPESLACLAGVAPSTRQSGKVKHVGFR-WAADKQLRDAVCDF
Mb2037|M.bovis_AF2122/97 GDCRARFPTPESLACLAGVAPSTRQSGKVKHVGFR-WAADKQLRDAVCDF
TH_3366|M.thermoresistible__bu GDSRAAYDGPDAVAALAGVTPVTKSSGKQHAVHFR-WACNKRFRRALTTF
Mflv_1197|M.gilvum_PYR-GCK GDDPNRYTTAKSRKNYAGTSPLTIASGKKRAVLAR-HVRNRRLYDAIDQW
Mvan_4491|M.vanbaalenii_PYR-1 GDDPNRYTTAKCRKNYAGTSPLTIASGRKRAVLAR-HVRNKRLYDALDQW
MSMEG_1732|M.smegmatis_MC2_155 GDDPNRYTDAKSRKNYAATSQITVASGRRRAVIAR-HIKNRHLADAIDMW
MAV_0496|M.avium_104 GDDRTRFADARGLKAFAGSAPITRASGKKTVVLHR-HIKNRRLAAVGPIW
MMAR_3782|M.marinum_M GGNLDGFDTVDRLACVAGLAPVPRDSGRISGNLHRPRRFNRRLLRPCYLA
*. : *. : . **: * ::::
Rv2014|M.tuberculosis_H37Rv AGDSRRANLWAADRYNRAIARGHDHPHAVRILARAWLYAIWHCWQDGAAY
Mb2037|M.bovis_AF2122/97 AGDSRRANLWAADRYNRAIARGHDHPHAVRILARAWLYAIWHCWQDGAAY
TH_3366|M.thermoresistible__bu ADNSRHQSTWAADIYNRARARGHDHPHAVRILARAWIRVIYRCWHDQSPY
Mflv_1197|M.gilvum_PYR-GCK AFCALTNSPGARTFYDQHRAAGDLHHQALRALGNRLVGILHGCLRHHTPY
Mvan_4491|M.vanbaalenii_PYR-1 AFCTLTRSPGARAYYDQHRAAGDTHHQALRALANRLVGILHGCLRHHSPY
MSMEG_1732|M.smegmatis_MC2_155 AYSSLRASPGARTFYDQHRAAGDTHHQALRALGNRFIGILHGCLRHHTHY
MAV_0496|M.avium_104 ALGSLRASPGARRHYDARRAAGDWNHQAQRHLFNKFLGQLHHCLQTGRLY
MMAR_3782|M.marinum_M ALSSLKNSPASRTFYDRKRAEGKSHKQALIALARRRINVIWAMLRDHTHY
* : . : *: * *. : :* * . : : : *
Rv2014|M.tuberculosis_H37Rv HPANHRAL-----------QALLNQDQDRAA------
Mb2037|M.bovis_AF2122/97 HPANHRAL-----------QALLNQDQDRAA------
TH_3366|M.thermoresistible__bu DPARHGAV-----------QKLHTAA-----------
Mflv_1197|M.gilvum_PYR-GCK SEHKAWAH-----------RQTDSRAA----------
Mvan_4491|M.vanbaalenii_PYR-1 DEHKAWAH-----------RKPAAA------------
MSMEG_1732|M.smegmatis_MC2_155 NEHTAWARGIVNTCGSGCFRRPPFRRADRRLRAMLAG
MAV_0496|M.avium_104 NEHLAFPLNRPGFDGGSCYWISTSGWSVRFVA-----
MMAR_3782|M.marinum_M HEPDLVNT-------------PLAA------------