For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
QCRAREERPGRKTDLLDAEWLVHLLECGLLRGWLIPPADIKAARDVIRYRRKLVEHRTSKLQRLGNVLQD AGIKADSVASSVTPKSVRAMVEALIDGERRPAVLADLARGSMRSKIPDLQRALEGRFDDHHALMCRLHLA HLDQLDAMIGALDEQIEQLMHPFCARRELIASIPGIGVGASATVISEIGADPAAWFPSAEHLASWVRLCP GNHESAGKRHHGARRTGNQHLQPVLVECAWAAVRTDGYLREYYRRQVRKFGGFRSPAANKKAITTVAHKL IVIIWHVLATGRPHQDLGADYFTTRMDPDKERRRLVAKLEAQGLGVTLEPAA*
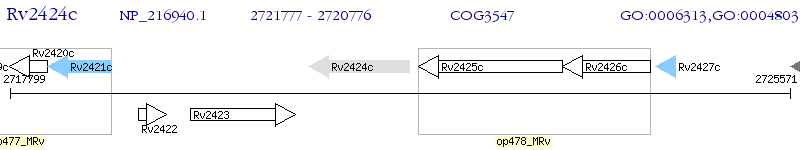
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv2424c | - | - | 100% (333) | PROBABLE TRANSPOSASE |
| M. tuberculosis H37Rv | Rv2177c | - | e-127 | 98.64% (221) | transposase |
| M. tuberculosis H37Rv | Rv2014 | - | 3e-08 | 26.42% (212) | transposase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2447c | - | 1e-132 | 98.70% (230) | transposase |
| M. gilvum PYR-GCK | Mflv_4200 | - | 4e-10 | 27.83% (230) | transposase IS116/IS110/IS902 family protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4550 | - | 2e-09 | 26.96% (230) | transposase |
| M. thermoresistible (build 8) | TH_3017 | - | 5e-07 | 28.95% (152) | PUTATIVE transposase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4882 | - | 8e-67 | 44.64% (336) | transposase IS116/IS110/IS902 family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4200|M.gilvum_PYR-GCK --------MTTGQVWAGVDVGKEHHWVCAVDDSGKVVLSRRLVNDEQPIR
MSMEG_4550|M.smegmatis_MC2_155 --------MAAGQVWAGVDVGKEHHWVCVVDDSGKVVLSRRLVNDEQPIR
TH_3017|M.thermoresistible__bu ---------------------------VVIDAQGKRLLSQRVANDEATLL
Rv2424c|M.tuberculosis_H37Rv --------------------------------------------------
Mb2447c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_4882|M.vanbaalenii_PYR-1 MLVESEDSEQIIARVAALDIGKAELVCCVRVPGRGTRRLQEVSTHSTMAR
Mflv_4200|M.gilvum_PYR-GCK ELVAEIDELAER--VSWTVDLTTVYAALVLTVLADAGKTVRYLAGRAVWQ
MSMEG_4550|M.smegmatis_MC2_155 ELVAEIDELAER--VLWAVDLTTVYAALLLTVLADAGKTVRYLAGRAVWQ
TH_3017|M.thermoresistible__bu KLISTVATMADGGEVTWAIDLNAGGAALLITLLIAADQRLLYIPGRTVHH
Rv2424c|M.tuberculosis_H37Rv ------------------------------------------MQCRAREE
Mb2447c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_4882|M.vanbaalenii_PYR-1 SLTELANHLVESGVQRVVMEATSDYWKPVFYLLEAHGLDPWLVNARDVKH
Mflv_4200|M.gilvum_PYR-GCK ASATYRGGEAKTDAKDARVIADQARMRGQDLPVLHPDDDLISELRMLTGH
MSMEG_4550|M.smegmatis_MC2_155 ASATYRGGEAKTDAKDARVIADQSRMRGQDLPVLHPNDDLISELRMLTGH
TH_3017|M.thermoresistible__bu ASGSYRG-EGKTDAKDAAVIADQARMR-RDLQPLRPGDDITVELRILTSR
Rv2424c|M.tuberculosis_H37Rv R-----PGR-KTDLLDAEWLVHLLECGLLRGWLIPPAD--IKAARDVIRY
Mb2447c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_4882|M.vanbaalenii_PYR-1 L-----PGRPKTDVLDAVWLAKLAERQMLRPSFVPPPP--IRQLRDLTRY
Mflv_4200|M.gilvum_PYR-GCK RADLVADRTRTINRLRQQLVAVCPALERAAQPSQDRGWVILLARYQRPKA
MSMEG_4550|M.smegmatis_MC2_155 RADLVADRTRTINRLRQQLVAVCPALERAAQLSQDRGWVVLLARYQRPKA
TH_3017|M.thermoresistible__bu RTDLVADRTRTINRLRAQLLEYFPALERAFDYSTSKAALLLLTGYQTPDG
Rv2424c|M.tuberculosis_H37Rv RRKLVEHRTSKLQRLGNVLQDAGIKADSVASSVTPKSVRAMVEAL-----
Mb2447c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_4882|M.vanbaalenii_PYR-1 RIDLVATRTAERNRVEKLLEDACIKLSVVASDIFGVSGRAMMAAL-----
Mflv_4200|M.gilvum_PYR-GCK IRQSGVSRLTKVLTDAGVRNAASIAAAAVAAVKTQTMRLPGEEVAAGLIA
MSMEG_4550|M.smegmatis_MC2_155 IRQSGVSRLTKVLTDAGVRNAASIAGAAVAAVKTQTVRLPGEEVAAGLVA
TH_3017|M.thermoresistible__bu LRRAGAARLAVWLGKRKARNADAVAAKALQAAHAQHTVIPGQQLAAAMVA
Rv2424c|M.tuberculosis_H37Rv IDGERRPAVLADLARGSMRSKIPDLQRALEGRFDDHHALMCR-LHLAHLD
Mb2447c|M.bovis_AF2122/97 --------MLADLARGSMRSKIPDLQRALEGRFDDHHALMCR-LHLAHLD
Mvan_4882|M.vanbaalenii_PYR-1 IAGERDPKVLAQLARSRMRTKIGRLEEAFNGHFNAHHGFLLT-RMLARLD
: * *. *. . : . :
Mflv_4200|M.gilvum_PYR-GCK DLAREVIALDDRIKTTDADIEGRFRRHPLAEVITSMPGIGFRLGAEFLAA
MSMEG_4550|M.smegmatis_MC2_155 DLAREVIALDDRIKTTDADIEGRFRRHPLAEVITSMPGMGFRLGAELLAA
TH_3017|M.thermoresistible__bu RLAKEVMALDTEIGDTDAMIEERFHRHRHAEIIVSMSGFGVTLGAEFLAA
Rv2424c|M.tuberculosis_H37Rv QLDAMIGALDEQIEQLMHPFCARR------ELIASIPGIGVGASATVISE
Mb2447c|M.bovis_AF2122/97 QLDAMIGALDEQIEQLMHPFCARR------ELIASIPGIGVGASATVISE
Mvan_4882|M.vanbaalenii_PYR-1 GIDADIAVLDEQIEVYLAPFAAAA------ARLDEIPGIGAVAAAIILAE
: : .** .* : : .:.*:* .* .::
Mflv_4200|M.gilvum_PYR-GCK VG-DPALIG-SADQLAAWAGLAPVSRDSGKRTGRLHTPKRYSRRLRRVMY
MSMEG_4550|M.smegmatis_MC2_155 VG-DPALIG-SADQLAAWAGLAPVSRDSRKRTGRLHTPKRYSRRLRRVMY
TH_3017|M.thermoresistible__bu TGGDMSAFD-SVDRLAGVAGLAPVPRDSGRISGNLQRPRRYNRRLLRVCY
Rv2424c|M.tuberculosis_H37Rv IGADPAAWFPSAEHLASWVRLCPGNHESAGKRHHGARR-TGNQHLQPVLV
Mb2447c|M.bovis_AF2122/97 IGADPAAWFPSAEHLASWVRLCPGNHESAGKRHHGARR-TGNQHLQPVLV
Mvan_4882|M.vanbaalenii_PYR-1 IGTD-MSVFPTAGHLCSWAKFSPGIQSSAGKTKGTASTGHGNRYLAQILG
* * :. :*.. . :.* :.* .: * :
Mflv_4200|M.gilvum_PYR-GCK MSALTAIRCDPDSKAYYQRKRDEGK------RPIPATICLARRRTNVLYA
MSMEG_4550|M.smegmatis_MC2_155 MSALTAIRCDPDSKAYYQRKRDEGK------RPIPATICLARRRTNVLYA
TH_3017|M.thermoresistible__bu LSALSSIRSDPASRTYYNRKRAEGK------RHSQAVLALARRRLNVLWA
Rv2424c|M.tuberculosis_H37Rv ECAWAAVRTDGYLREYYRRQVRKFGGFRSPAANKKAITTVAHKLIVIIWH
Mb2447c|M.bovis_AF2122/97 ECAWAAVRTDGYLREYYRRQVRKFGGFRSPAANKKAIIAVAHKLIVIIWH
Mvan_4882|M.vanbaalenii_PYR-1 EAAIAASRTDTFLGQRYRRIARRRG-------NKRALVAVGRSILVIIWH
.* :: * * *.* . * :.: :::
Mflv_4200|M.gilvum_PYR-GCK LIRD-------------NRTWQPDSPPVTAAA---------------
MSMEG_4550|M.smegmatis_MC2_155 LIRD-------------NRTWQPESPPNIAAA---------------
TH_3017|M.thermoresistible__bu MLRD-------------HTTYQPTTPTAAAAA---------------
Rv2424c|M.tuberculosis_H37Rv VLAT-GRPHQDLGADYFTTRMDPDKERRRLVAKLEAQGLGVTLEPAA
Mb2447c|M.bovis_AF2122/97 VLAT-GRPYQDLGADYFTTRMDPDKERRRLVAKLEAQGLGVTLEPAA
Mvan_4882|M.vanbaalenii_PYR-1 LLTDPDATFIDLGPDHYTKHLNPETKRRNHIRQLEALGYTVTLTPAA
:: :* .