For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LTLFIGDDWAEDHHDIHLMDAEGTRLASRRLPEGLAGIRGFHELVAAHAQDPTEVVIGIETDRGLWVEAL AAAGYEVFAVNPLAVARYRDRHQVSGAKSDAADAKLLADLVRTDRHNHRPIAGDTPEVEAIKVLARAHQN LIWSRNRNTNALRSALREYYPAALEAFASLSDRDALAILGRAPTPTDAARLSVSKIRSALKAAGRQRNLD AKALEIQTALRSDQLAAAPAVTAAFGATTRAAVAIITELNHHIADLEAELAAHFETHPDADIYLSLPGLG VILGARVLGEFGDDPNRYTTAKSRKNYAGTSPLTIASGKKRAVLARHVRNRRLYDAIDQWAFCALTNSPG ARTFYDQHRAAGDLHHQALRALGNRLVGILHGCLRHHTPYSEHKAWAHRQTDSRAA
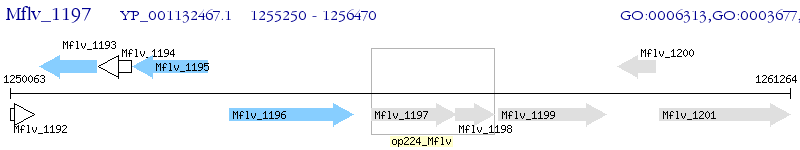
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1197 | - | - | 100% (406) | transposase, IS111A/IS1328/IS1533 |
| M. gilvum PYR-GCK | Mflv_4200 | - | 5e-25 | 27.08% (384) | transposase IS116/IS110/IS902 family protein |
| M. gilvum PYR-GCK | Mflv_1737 | - | 5e-25 | 27.08% (384) | transposase IS116/IS110/IS902 family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2036 | - | 2e-17 | 32.12% (193) | transposase |
| M. tuberculosis H37Rv | Rv2014 | - | 2e-15 | 32.78% (180) | transposase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_3782 | - | 4e-36 | 29.55% (396) | transposase |
| M. avium 104 | MAV_0496 | - | 5e-85 | 45.62% (377) | transposase IS116/IS110/IS902 |
| M. smegmatis MC2 155 | MSMEG_4081 | - | 0.0 | 82.62% (397) | transposase IS116/IS110/IS902 |
| M. thermoresistible (build 8) | TH_2233 | - | 0.0 | 85.64% (390) | Transposase IS116/IS110/IS902 family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4491 | - | 0.0 | 78.39% (398) | transposase, IS111A/IS1328/IS1533 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1197|M.gilvum_PYR-GCK -MTLFIGDDWAEDHHDIHLMDAEGTRLASRRLPEGLAGIRGFHELVAAHA
TH_2233|M.thermoresistible__bu -------------------MDAEGAKLASRRLPEGLAGIGGFHDLVAAHA
MSMEG_4081|M.smegmatis_MC2_155 --MLFVGDDWAEDHHDLYLMNEAGDRLASRRLPEGLAGIRLLHDLIAAHA
Mvan_4491|M.vanbaalenii_PYR-1 --MIFVGDDWAEDHHDVYLMDESGQRLAARRLPEGLSGIRALHELIAGYA
MAV_0496|M.avium_104 -------------------MVARG------RVSNDAAGFAALLTLLAEAG
Rv2014|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3782|M.marinum_M MPTVWAGVDTGKRTHHCVVLDRNGGVLLSRPVDNDETALLELIEAVVGVA
Mb2036|M.bovis_AF2122/97 ----------------MSIVDARGREVRRATIEHNAAGLRELLELLSRAG
Mflv_1197|M.gilvum_PYR-GCK QDPTE-VVIGIETDRGLWVEALAAAGYEVFAVNPLAVARYRDRHQVSGAK
TH_2233|M.thermoresistible__bu EEPAQ-VVIGIETDRGLWVEALAAAGYQVFAINPLAVARYRDRHQVSGAK
MSMEG_4081|M.smegmatis_MC2_155 DDPAQ-VAVGIETDRGLWVEALTGAGYQVYAVNPLAVARYRDRHAVSGAK
Mvan_4491|M.vanbaalenii_PYR-1 EEPDQ-VIVGIETDRGLWVSALAAAGYRVWAINPMAAARYRDRHHVSGAK
MAV_0496|M.avium_104 DSAAHPIPVGIETDRGLWVAALRETGRVIYPINPLAASRYRARHAVSGAK
Rv2014|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3782|M.marinum_M AGREVCWATDLNAGGAAMLITLLAAHDQQLLYIPGRIVHHAAATYRGDGK
Mb2036|M.bovis_AF2122/97 -----AREVAIERPDGPVVDTLLEAGITVVVISPNQLKNLRGRYGSAGNK
Mflv_1197|M.gilvum_PYR-GCK SDAADAKLLADLVRTDRHNHRPIAGDTPEVEAIKVLARAHQNLIWSRNRN
TH_2233|M.thermoresistible__bu SDAGDAKVLADLVRTDRHNHRPIAGDTPQVEAIKVLARAHQSLIWARTRH
MSMEG_4081|M.smegmatis_MC2_155 SDAADAKLLADLVRTDRHNHRLIAGDTPDAEAVKVLARAHQNLIWTRNRH
Mvan_4491|M.vanbaalenii_PYR-1 SDASDAKLLADLVRTDRHNHREIAGDRPQAEAIKVLARSHQSLIWARTRH
MAV_0496|M.avium_104 SDATDAVLLANIVRTDAAAHRPLPADTELAQAIRVLARAQQDAVWARQQI
Rv2014|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3782|M.marinum_M TDAKNARIIADQAR-MRTDLQPVRRADPIATDLRLLTSRRTDVVCDRVRA
Mb2036|M.bovis_AF2122/97 DDRFDAFVLADTLRTDRSRLRPLLPDTPATATLRRTCRPRKDLVAHRVAL
Mflv_1197|M.gilvum_PYR-GCK TNALRSALREYYPAALEAFASLSD-----RDALAILGRAPTPTDAARLSV
TH_2233|M.thermoresistible__bu TNTLRSALREYYPAALEAFDDLAD-----RDALAILGRAPTPTDATGLSL
MSMEG_4081|M.smegmatis_MC2_155 TNALRSALREYYPAALEAFDDLSD-----RDALAILGHAPDPRQASSLSL
Mvan_4491|M.vanbaalenii_PYR-1 ANMLRSGLREYYPAALEAFDSLTD-----SDALAILGRAPTPDQGARLSL
MAV_0496|M.avium_104 GNQIRDLLKDFYPAAIAAFAGLPEGGLARADARTILAAAPTPTQAATLTP
Rv2014|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_3782|M.marinum_M INRLRQTLLEYFPPLERAFDYSKS-----KAALTLLSRYQTPDSLRRIGP
Mb2036|M.bovis_AF2122/97 ANQLRAHLRVVFPG------------------------------------
Mflv_1197|M.gilvum_PYR-GCK SKIRSALKAAGRQRNLDAKALEIQTALRSDQLAAAPAVTAAFGATTRAAV
TH_2233|M.thermoresistible__bu PKIRSALKTAGRQRNLDARALQIQTALRTEQLTAPASVTAAFGATTRATV
MSMEG_4081|M.smegmatis_MC2_155 AKIRSALKAAGRQRNIDIRAQEIQAALRSEQLAAPAAVTAAFAATTRATV
Mvan_4491|M.vanbaalenii_PYR-1 AKIGSALKSAGRQRNIESRAAEIQTALRGEHLTAPPAVAAAFGATTAAAV
MAV_0496|M.avium_104 ARLRRLLTKAGRRRHLDRDIERLRAVFTDTYLHQPPAVENAMGIQLAALL
Rv2014|M.tuberculosis_H37Rv ---------------------MLHDRLTGAPRGATGDEGAANAHITRAMV
MMAR_3782|M.marinum_M TRLATWLKARGCRNSATVAQTAVQAAQTQHTTLPTQTVGAQLVPR---LA
Mb2036|M.bovis_AF2122/97 ------------------------------------------------VV
Mflv_1197|M.gilvum_PYR-GCK AIITELNHHIADLEAELAAHFETHPDADIYLSLPGLGVILGARVLGEFGD
TH_2233|M.thermoresistible__bu GIITELNRQIDNLDAELTSHFEMHPDADIYRSLPGLGVILGARVLGEFGD
MSMEG_4081|M.smegmatis_MC2_155 GLVVELSRQIDDLENELAAHFETHPDADIYRSLPGLGVILGARVLGEFGD
Mvan_4491|M.vanbaalenii_PYR-1 HVITALNDQIAELETALADHFEAHPDADIYRSLPGLGVILGARVLGEFGD
MAV_0496|M.avium_104 RQLEAACTAADELAEAAIAHFEQHPDAKIITSFPGLGMLAGARVLAEIGD
Rv2014|M.tuberculosis_H37Rv AALTSVATQIKTLDAQIAEQLSLHADAHIFTSLPRSGTVRAARLLAEIGD
MMAR_3782|M.marinum_M AEITTIDAELAHVDNEITARFADHDSAEILTSMPGFGPVLAATFLAQIGG
Mb2036|M.bovis_AF2122/97 GLFADLDSPISLAFLTFLPRFDCQDRADWLSVKRLAGWLAAAGYCGRAPR
. :: : *. * : .* ..
Mflv_1197|M.gilvum_PYR-GCK DPNRYTTAKSRKNYAGTSPLTIASGKKRAVLAR-HVRNRRLYDAIDQWAF
TH_2233|M.thermoresistible__bu DPNRYTSAKSRKNYAGTSPLTIASGKKRAVLAR-HVRNRRLYDAIDQWAF
MSMEG_4081|M.smegmatis_MC2_155 DPNRYTNAKCRKNYAGTSPLTIASGRKRAVLAR-HIRNRRLYDAIDQWAF
Mvan_4491|M.vanbaalenii_PYR-1 DPNRYTTAKCRKNYAGTSPLTIASGRKRAVLAR-HVRNKRLYDALDQWAF
MAV_0496|M.avium_104 DRTRFADARGLKAFAGSAPITRASGKKTVVLHR-HIKNRRLAAVGPIWAL
Rv2014|M.tuberculosis_H37Rv CRARFPTPESLACLAGVAPSTRQSGKVKHVGFR-WAADKQLRDAVCDFAG
MMAR_3782|M.marinum_M NLDGFDTVDRLACVAGLAPVPRDSGRISGNLHRPRRFNRRLLRPCYLAAL
Mb2036|M.bovis_AF2122/97 PAHRCPARRHR---------------------------------------
Mflv_1197|M.gilvum_PYR-GCK CALTNSPGARTFYDQHRAAGDLHHQALRALGNRLVGILHGCLRHHTPYSE
TH_2233|M.thermoresistible__bu CALTNSPGARAFYDQRRAADDLHHQALRALGNRLVGILHGCLRHHTRYDE
MSMEG_4081|M.smegmatis_MC2_155 CALTRSPGARQFYDHRRAEGDLHHQALRALGNRLVGILHGCLRHRTRYDE
Mvan_4491|M.vanbaalenii_PYR-1 CTLTRSPGARAYYDQHRAAGDTHHQALRALANRLVGILHGCLRHHSPYDE
MAV_0496|M.avium_104 GSLRASPGARRHYDARRAAGDWNHQAQRHLFNKFLGQLHHCLQTGRLYNE
Rv2014|M.tuberculosis_H37Rv DSRRANLWAADRYNRAIARGHDHPHAVRILARAWLYAIWHCWQDGAAYHP
MMAR_3782|M.marinum_M SSLKNSPASRTFYDRKRAEGKSHKQALIALARRRINVIWAMLRDHTHYHE
Mb2036|M.bovis_AF2122/97 --------------------------------------------------
Mflv_1197|M.gilvum_PYR-GCK HKAWAHRQT--DSRAA--------------
TH_2233|M.thermoresistible__bu HTAWAHRQTTADTQAA--------------
MSMEG_4081|M.smegmatis_MC2_155 HKAWAHRTP----AAA--------------
Mvan_4491|M.vanbaalenii_PYR-1 HKAWAHRKP----AAA--------------
MAV_0496|M.avium_104 HLAFPLNRPGFDGGSCYWISTSGWSVRFVA
Rv2014|M.tuberculosis_H37Rv ANHRALQALLNQDQDRAA------------
MMAR_3782|M.marinum_M PDLVNTPLAA--------------------
Mb2036|M.bovis_AF2122/97 ------------------------------