For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGEANIREQAIATMPRGGPDASWLDRRFQTDALEYLDRDDVPDEVKQKIIGVLDRVGTLTNLHEKYARIA LKLVSDIPNPRILELGAGHGKLSAKILELHPTATVTISDLDPTSVANIAAGELGTHPRARTQVIDATAID GHDHSYDLAVFALAFHHLPPTVACKAIAEATRVGKRFLIIDLKRQKPLSFTLSSVLLLPLHLLLLPWSSM RSSMHDGFISALRAYSPSALQTLARAADPGMQVEILPAPTRLFPPSLAVVFSRSSSAPTESSECSADRQP GE
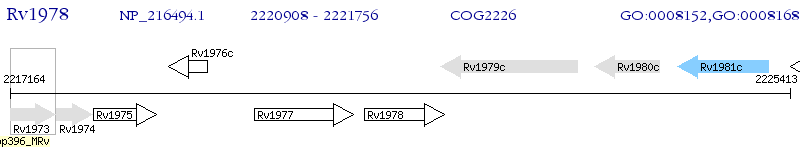
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1978 | - | - | 100% (282) | hypothetical protein Rv1978 |
| M. tuberculosis H37Rv | Rv3038c | - | 3e-05 | 27.27% (110) | hypothetical protein Rv3038c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2000 | - | 1e-159 | 99.65% (282) | hypothetical protein Mb2000 |
| M. gilvum PYR-GCK | Mflv_1326 | - | 3e-59 | 47.86% (257) | methyltransferase type 11 |
| M. leprae Br4923 | MLBr_01723 | - | 4e-05 | 26.36% (110) | hypothetical protein MLBr_01723 |
| M. abscessus ATCC 19977 | MAB_4700c | - | 7e-66 | 54.17% (240) | hypothetical protein MAB_4700c |
| M. marinum M | MMAR_1415 | - | 3e-08 | 34.02% (97) | hypothetical protein MMAR_1415 |
| M. avium 104 | MAV_0081 | - | 4e-89 | 64.15% (265) | hypothetical protein MAV_0081 |
| M. smegmatis MC2 155 | MSMEG_6231 | - | 1e-61 | 51.90% (237) | hypothetical protein MSMEG_6231 |
| M. thermoresistible (build 8) | TH_2189 | - | 8e-05 | 28.30% (106) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0820 | - | 5e-06 | 32.38% (105) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_5469 | - | 8e-67 | 52.33% (258) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv1978|M.tuberculosis_H37Rv --------------------------------------------------
Mb2000|M.bovis_AF2122/97 --------------------------------------------------
MAV_0081|M.avium_104 --------------------------------------------------
Mflv_1326|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5469|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6231|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4700c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
TH_2189|M.thermoresistible__bu VIRRLMATALTALTTAVLAGCGDAGSWVEAKPAEGWSAQYGDAANSSYTS
MMAR_1415|M.marinum_M --------------------------------------------------
MUL_0820|M.ulcerans_Agy99 --------------------------------------------------
Rv1978|M.tuberculosis_H37Rv --------------------------------------------------
Mb2000|M.bovis_AF2122/97 --------------------------------------------------
MAV_0081|M.avium_104 --------------------------------------------------
Mflv_1326|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5469|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6231|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4700c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
TH_2189|M.thermoresistible__bu AAGAEALTLEWSRSVKGELAAQVAVGASGYLAVNAQTPAGCSLMVWEYAN
MMAR_1415|M.marinum_M --------------------------------------------------
MUL_0820|M.ulcerans_Agy99 --------------------------------------------------
Rv1978|M.tuberculosis_H37Rv --------------------------------------------------
Mb2000|M.bovis_AF2122/97 --------------------------------------------------
MAV_0081|M.avium_104 --------------------------------------------------
Mflv_1326|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5469|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6231|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4700c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
TH_2189|M.thermoresistible__bu SARQRWCTRLVQGGGRTSPLLDGFDNVYIGQPGAILSFPPTQWIRWRKPV
MMAR_1415|M.marinum_M --------------------------------------------------
MUL_0820|M.ulcerans_Agy99 --------------------------------------------------
Rv1978|M.tuberculosis_H37Rv --------------------------------------------------
Mb2000|M.bovis_AF2122/97 --------------------------------------------------
MAV_0081|M.avium_104 --------------------------------------------------
Mflv_1326|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5469|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6231|M.smegmatis_MC2_155 --------------------------------------------------
MAB_4700c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01723|M.leprae_Br4923 --------------------------------------------------
TH_2189|M.thermoresistible__bu IGMPTTPRILAPGELLVVTHLGQVLLFDAHRGTVTGTPLDLVAGVDPTDS
MMAR_1415|M.marinum_M --------------------------------------------------
MUL_0820|M.ulcerans_Agy99 --------------------------------------------------
Rv1978|M.tuberculosis_H37Rv ------------------------------------------MGEANIRE
Mb2000|M.bovis_AF2122/97 ------------------------------------------MGEANIRE
MAV_0081|M.avium_104 ------------------------------------------MSEALTVD
Mflv_1326|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5469|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6231|M.smegmatis_MC2_155 ------------------------------------------MAYMATST
MAB_4700c|M.abscessus_ATCC_199 ------------------------------------------MFYVENSA
MLBr_01723|M.leprae_Br4923 ------------------------------------------MTRSTEIS
TH_2189|M.thermoresistible__bu ERGLADCAGARXXXCWPPPRPPSTVTPRARDAARHPTARLPAMTETTEPG
MMAR_1415|M.marinum_M --------------------------------------------------
MUL_0820|M.ulcerans_Agy99 --------------------------------------------------
Rv1978|M.tuberculosis_H37Rv QAIA-----TMPRGGPDASWLDRRFQTDALEYLDRDDVP----DEVKQKI
Mb2000|M.bovis_AF2122/97 QAIA-----TMPRGGPDASWLDRRFQTDALEYLDRDDVP----DEVKQKI
MAV_0081|M.avium_104 PAIA-----SMPRGGPDASWLDRRLQTEKLEYTDRYDIP----DEVKQTV
Mflv_1326|M.gilvum_PYR-GCK ----------MPRGGPRASCLDRLLETDRQEYLDRASELPADVAR-KRSV
Mvan_5469|M.vanbaalenii_PYR-1 ----------MPRGGPDASCLDRLLQTDRQEYLDRDSAGDAGLEQTKRSV
MSMEG_6231|M.smegmatis_MC2_155 ADIA-----AMPRGGPRASCLDRLLQTDRLEYLDRDDVD----QRVKRSV
MAB_4700c|M.abscessus_ATCC_199 RDIS-----SMPRGGPNASWLDRRLQTDRLEYLDRDDVD-----DLKRRV
MLBr_01723|M.leprae_Br4923 AN-------AVPNPHATAEQVAAARKDSKLAQVLYHDWEAEN-YDEKWSI
TH_2189|M.thermoresistible__bu IGSEGAGDLPAPNPHATAEQVEAAMRDSKLAQILYHDWEAES-YDDKWSI
MMAR_1415|M.marinum_M --------------MPAMSRIESVFCRSAP--------------------
MUL_0820|M.ulcerans_Agy99 ----------MTNHHPRFGLQHSGESRDYLPAAGHDALLP----------
. .
Rv1978|M.tuberculosis_H37Rv IGVLDRVG--TLTNLHEKYARIALKLVSDIPNPRILELGAGHGKLSAKIL
Mb2000|M.bovis_AF2122/97 IGVLDRVG--TLTNLHEKYARIALKLVSDIPNPRILELGAGHGKLSAKIL
MAV_0081|M.avium_104 VAALDRMG--TRAGYHERNARTALDIVADIANPRILELGAGHGKLSAEIL
Mflv_1326|M.gilvum_PYR-GCK IRALEWTG--ETFGHHVRFAEIVLDQISDVPDPKILELGAGHGALSRKLL
Mvan_5469|M.vanbaalenii_PYR-1 IRALDWTG--EVFGNHEKFAKIALDEVADVADPRILELGAGHGGLSRKLL
MSMEG_6231|M.smegmatis_MC2_155 VRALEWTG--DMFGNHEKFAAISLDLLAEVPDPRILELGAGHGSVSRRLL
MAB_4700c|M.abscessus_ATCC_199 MQSLDRAGRRRWIGIHEKCARMAIREVADVDRPKILELGAGLGGLSHKLL
MLBr_01723|M.leprae_Br4923 SYDQRCVD----YVRHCFDAIVPDELFTELPYDCALELGCGTGFFLLNLI
TH_2189|M.thermoresistible__bu SYDQRCVD----YARGRFDAIVPDHVQRELPYDNALELGCGTGFFLLNLI
MMAR_1415|M.marinum_M ------------WRAATRRLVLPWALQGIEPVGEVLEIGAGSGAMAAEML
MUL_0820|M.ulcerans_Agy99 --AYDLLS---RLLGMNKIHRTLITQAELADCGRILEIGCGTGNLAIKAK
**:*.* * . .
Rv1978|M.tuberculosis_H37Rv ELHPTATVTISDLDPTSVANIAAGELGTHPRARTQVIDATAIDGHDHSYD
Mb2000|M.bovis_AF2122/97 ELHPTATVTISDLDPTSVANIAAGELGTHPRARTQVIDATAIDGHGHSYD
MAV_0081|M.avium_104 TSHPTATVTVSDLDPTSVANIAAGPLGADPRARTQVVDATAIDAPDDSYD
Mflv_1326|M.gilvum_PYR-GCK DWHPTARLTVTDIEPASVAAIADSDVGAHPRATVRQMDATALDAADGEFD
Mvan_5469|M.vanbaalenii_PYR-1 EWHPSARLTVTDVEPASVAAIAAGDLGSHPRATVRQMDATALDAADGEFD
MSMEG_6231|M.smegmatis_MC2_155 ANHPTAHVTVTDVDPAAVAAMADGELGDHPRAVVRAMDATDIDADDGSFD
MAB_4700c|M.abscessus_ATCC_199 EMHSTAQVTVTDVDPSFVAAVAAGEIGSHPRATVRQLDATEIDSPGGYYD
MLBr_01723|M.leprae_Br4923 QSGVARRGSVTDLSPGMVKIATRNGQSLGLDINGRVADAEGIPYDDNTFD
TH_2189|M.thermoresistible__bu QSGVARRGSVTDLSPGMVRVATRNGRSLGLDVDGRVADAEGIPYDDNTFD
MMAR_1415|M.marinum_M AAQPDTRITVTDYDDAMVAAAAQRLAPFQTRATARQADATALPFGDNTFD
MUL_0820|M.ulcerans_Agy99 RAQPRAEVIGSDPDPRALQRAQR-KTGNVEEVRFDQGYAQRLPYADGEFD
:* . : * : . :*
Rv1978|M.tuberculosis_H37Rv LAVFALAFHHLP---PTVACKAIAEATRVGKRFLIIDLKRQKPLSFTLSS
Mb2000|M.bovis_AF2122/97 LAVFALAFHHLP---PTVACKAIAEATRVGKRFLIIDLKRQKPLSFTLSS
MAV_0081|M.avium_104 LVVFAQAFHHLP---PATAVRAIAEATRVGKRFLVIDLPRPRSVQLLLTP
Mflv_1326|M.gilvum_PYR-GCK LAVFALSFHHLP---PAAAARVIAEGTRAADTLLIIDLPRP--------P
Mvan_5469|M.vanbaalenii_PYR-1 LAVFALSFHHLP---PQLAARAIAEGARVADKLLIIDLPRP--------P
MSMEG_6231|M.smegmatis_MC2_155 LALFALSFHHLS---PAQAAAVFAEGTRVARTLLIIDLPRP--------P
MAB_4700c|M.abscessus_ATCC_199 LAVFALSLHHLP---PELAARVFATGTRVARKLLIVDLPRP--------P
MLBr_01723|M.leprae_Br4923 LVVGHAVLHHIPD--VELSLREVLRVLKPGGRFVFAGEPTDVGNRYARVF
TH_2189|M.thermoresistible__bu LVVGHAVLHHIPD--VELSLREVVRVLRPGGRFVFAGEPTTVGNSYARAL
MMAR_1415|M.marinum_M TVLTFIMLHHTVR--WEQTLAEAVRVLAPHGTLIGYDLLSS---------
MUL_0820|M.ulcerans_Agy99 RVLSSMMLHHLDEDAKSAAAAEAFRVLRPGGRLHLVDVGGD---------
.: :** : : .
Rv1978|M.tuberculosis_H37Rv VLLLPLH-LLLLPWSSMRSSMHD-------------GFISALRAYSPSAL
Mb2000|M.bovis_AF2122/97 VLLLPLH-LLLLPWSSMRSSMHD-------------GFISALRAYSPSAL
MAV_0081|M.avium_104 LILAPFAAALALVRPSLKHVMHD-------------AYISALRAYSRSAF
Mflv_1326|M.gilvum_PYR-GCK APLHLVRLATMLPWAPLVPFVHD-------------GVISSLRAYSPAAL
Mvan_5469|M.vanbaalenii_PYR-1 APLHVLRLAAMLPWAPLVPFLHD-------------GVISSLRAYSPSAL
MSMEG_6231|M.smegmatis_MC2_155 APLHLLRLATMLPLAPLVPFVHD-------------GVISSLRCYSPSAL
MAB_4700c|M.abscessus_ATCC_199 VPLHLALLAVVLPLVAISPLQHD-------------GFISSLRAYSPSAL
MLBr_01723|M.leprae_Br4923 ADLTWKVTIRVVQLPGLSAWRRTQVEIDENSRAAALEWVVDLHTFEPKVL
TH_2189|M.thermoresistible__bu STLTWHVATTVTRLPGLQGWRRPQAELDESSRAAALEAVVDLHTFEPSDL
MMAR_1415|M.marinum_M --------WPTRTLHRLEGAPHR---------------LIERGELQP-VL
MUL_0820|M.ulcerans_Agy99 --------MSAHDGLASRFIRHN---------------LHVAGNLGDGIR
: :
Rv1978|M.tuberculosis_H37Rv QTLARAADPGMQVEILP-APTRLFPPSLAVVFSRSSSAPTESSECSADRQ
Mb2000|M.bovis_AF2122/97 QTLARAADPGMQVEILP-APTRLFPPSLAVVFSRSSSAPTESSECSADRQ
MAV_0081|M.avium_104 IALGKAADPRMGVEFLPLSACRLRPGHVGIVFTRPRAS------------
Mflv_1326|M.gilvum_PYR-GCK RALAAHAGEDIDIELRG---G-LFSPQVAVAARR----------------
Mvan_5469|M.vanbaalenii_PYR-1 RALATHADPAVDIELRG---G-LLSPQVAVATRR----------------
MSMEG_6231|M.smegmatis_MC2_155 RALAAHAG--VALELRG---G-LTTPQIAIARRA----------------
MAB_4700c|M.abscessus_ATCC_199 RALAHHADPAINVEFRS---RRGLGPTVVVASRP----------------
MLBr_01723|M.leprae_Br4923 ETMATNAGAVQVKTVTEEFTAAMLGWPLRTFESTMPPSKLGWGWARFAFT
TH_2189|M.thermoresistible__bu ERMARNAGAVEVATASEEFTAAMLGWPVRTFEAAVPPGKLGWNWAKFAFG
MMAR_1415|M.marinum_M RQLPLSASRVRQNRLLVRFTAMKQASPS----------------------
MUL_0820|M.ulcerans_Agy99 RLLDTAGFDVAEVGAQR---HPLLGRLVYYRATRPCA-------------
: .
Rv1978|M.tuberculosis_H37Rv PGE------------------------------
Mb2000|M.bovis_AF2122/97 PGE------------------------------
MAV_0081|M.avium_104 ---------------------------------
Mflv_1326|M.gilvum_PYR-GCK ---------------------------------
Mvan_5469|M.vanbaalenii_PYR-1 ---------------------------------
MSMEG_6231|M.smegmatis_MC2_155 ---------------------------------
MAB_4700c|M.abscessus_ATCC_199 ---------------------------------
MLBr_01723|M.leprae_Br4923 SWKTLSWVDANVWRRVIPKSWFYNIIITGVKPS
TH_2189|M.thermoresistible__bu SWTALSWVDANVWRRVMPKGWFYNVMVTGVKPS
MMAR_1415|M.marinum_M ---------------------------------
MUL_0820|M.ulcerans_Agy99 ---------------------------------