For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDILRTPDERFADLPDFPFAPHYVEVDGLRMHYLDEGPADGPVVLLLHGEPSWSYLYRRMIPVLVDAGLR AVAIDLVGFGRSDKPTRREDYTYQAHVDWTWAAIEAIGLTGITLVCQDWGGLIGLRLVGEHPDRFARVVA ANTTLPTGDQRPGEAFLAWQRFSQETPDFAVGHIVNGGCVSTLAPEVIAAYDAPFPDDSYKAGARQFPTL VPTSPDDPAAPANRAAWEVLRTFDRPFLCAFSDSDPITRGADAPLRKFIPGAQGRPEVTIAGGGHFLQED KGPELAAVVVDFIAANPV
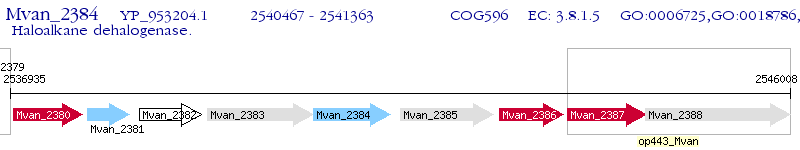
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2384 | - | - | 100% (298) | haloalkane dehalogenase |
| M. vanbaalenii PYR-1 | Mvan_4733 | - | 1e-16 | 27.12% (295) | alpha/beta hydrolase fold |
| M. vanbaalenii PYR-1 | Mvan_3998 | - | 2e-15 | 34.29% (140) | alpha/beta hydrolase fold |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2318 | - | 8e-93 | 57.39% (291) | haloalkane dehalogenase |
| M. gilvum PYR-GCK | Mflv_4004 | - | 8e-56 | 83.90% (118) | haloalkane dehalogenase |
| M. tuberculosis H37Rv | Rv2296 | - | 8e-93 | 57.39% (291) | haloalkane dehalogenase |
| M. leprae Br4923 | MLBr_02297 | - | 9e-09 | 31.03% (145) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_0528c | - | 7e-14 | 29.46% (129) | epoxide hydrolase EphA |
| M. marinum M | MMAR_3472 | - | 9e-91 | 54.55% (297) | haloalkane dehalogenase |
| M. avium 104 | MAV_2130 | - | 5e-94 | 55.93% (295) | haloalkane dehalogenase |
| M. smegmatis MC2 155 | MSMEG_6314 | - | 5e-94 | 57.86% (299) | haloalkane dehalogenase |
| M. thermoresistible (build 8) | TH_1615 | linB | 1e-24 | 31.74% (293) | POSSIBLE HALOALKANE DEHALOGENASE DHAA (1-CHLOROHEXANE |
| M. ulcerans Agy99 | MUL_3052 | - | 4e-20 | 27.49% (291) | haloalkane dehalogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2384|M.vanbaalenii_PYR-1 ---------MDILRTPDERFADLPDFPFAPHYVEVDG-----LRMHYLDE
Mflv_4004|M.gilvum_PYR-GCK ---------MDVLRTPYERFTDLPGFPFDPHYVEVGG-----LRMHYLDE
MSMEG_6314|M.smegmatis_MC2_155 ---------MQTLRTPDERFAQLPEFPYAPMYCDIDDGEGGRLRVAWVED
Mb2318|M.bovis_AF2122/97 ---------MDVLRTPDSRFEHLVGYPFAPHYVDVTAGDTQPLRMHYVDE
Rv2296|M.tuberculosis_H37Rv ---------MDVLRTPDSRFEHLVGYPFAPHYVDVTAGDTQPLRMHYVDE
MAV_2130|M.avium_104 MPVHAEELNMHVLRTPDSRFENLEDYPFVAHYLDVTARDTRPLRMHYLDE
MMAR_3472|M.marinum_M ---------MDVVRTPDARFQNLVGYPFAAHYVDVAATDTPHLRMHYIDE
MUL_3052|M.ulcerans_Agy99 ---------MGRQFTPD-FTPDPKLYPFASRWFDSSRG-----RLHYIDE
TH_1615|M.thermoresistible__bu -------------------------MPGSKPYGEPRFREIKNKRMAYIDE
MAB_0528c|M.abscessus_ATCC_199 -------------------------MTDAAVDIRERDIATNGVHLRIVEA
MLBr_02297|M.leprae_Br4923 ------MAMSNPSVTRIAGPWHHLDVHANGIRFHVVEAVPTQPAGQDCAP
Mvan_2384|M.vanbaalenii_PYR-1 GPADGPVVLLLHGEPSWSYLYRRMIPVLVDAGLRAVAIDLVGFGRSD--K
Mflv_4004|M.gilvum_PYR-GCK GPAGGDVVLLLHGEPSWSYLYRTMIPVLVDAGLRAVAIDLVGFGRSD--K
MSMEG_6314|M.smegmatis_MC2_155 GPAHAEPVLLLHGEPSWSFLYRKMIPILVAAGHRVICPDLVGFGRSD--K
Mb2318|M.bovis_AF2122/97 GPGDGPPIVLLHGEPTWSYLYRTMIPPLSAAGHRVLAPDLIGFGRSD--K
Rv2296|M.tuberculosis_H37Rv GPGDGPPIVLLHGEPTWSYLYRTMIPPLSAAGHRVLAPDLIGFGRSD--K
MAV_2130|M.avium_104 GPIDGPPIVLLHGEPTWSYLYRTMITPLTDAGNRVLAPDLIGFGRSD--K
MMAR_3472|M.marinum_M GPADGPPIVLLHGEPTWSYLYRTMIPPLAAGGYRVLAPDLIGFGRSD--K
MUL_3052|M.ulcerans_Agy99 --SDGPPILLCHGNPTWSFLYRHIIVALRDH-FRCIAPDYLGFGLSE--H
TH_1615|M.thermoresistible__bu G--EGDAIVFQHGNPTSSYLWRNVLP-HLEGLGRLVACDLIGMGGSEKLS
MAB_0528c|M.abscessus_ATCC_199 GEPGQPAVILSHGFPELAYSWRHQIPALAAAGYHVIAPDQRGYGRSS--M
MLBr_02297|M.leprae_Br4923 SATARPLVMLLHGFGSFWWSWRHQLRGLTEA--RLVAVDLRGYGGSDK-P
::: ** : :* : : :. * * * *.
Mvan_2384|M.vanbaalenii_PYR-1 PTRREDYTYQAHVDWTWA-AIEAIGLTGITLVCQDWGGLIGLRLVGEHPD
Mflv_4004|M.gilvum_PYR-GCK PADRADYTYQAHVDWTWG-AVAALGLADITLVCQD---------------
MSMEG_6314|M.smegmatis_MC2_155 PTRIEDHTYARHVEWMRSLVFDALDLRKVTLVGQDWGGLIGLRLAAEHPD
Mb2318|M.bovis_AF2122/97 PTRIEDYTYLRHVEWVTS-WFENLDLHDVTLFVQDWGSLIGLRIAAEHGD
Rv2296|M.tuberculosis_H37Rv PTRIEDYTYLRHVEWVTS-WFENLDLHDVTLFVQDWGSLIGLRIAAEHGD
MAV_2130|M.avium_104 PSRIEDYSYQRHVDWVVS-WFEHLNLSDVTLFVQDWGSLIGLRIAAEQPD
MMAR_3472|M.marinum_M PTRIADYTYLRHVEWVKS-WFEELRLAEATLFVQDWGSLIGLRVAAEHGD
MUL_3052|M.ulcerans_Agy99 PGTFG-YKVDEHAQVVGE-FVDHLGLDNYLTMGQDWGGPISMAVAVERAE
TH_1615|M.thermoresistible__bu PSGPDRYHYGEHRDYLFALWEELELGDRVVLVLHDWGSVLGFDWANHHRD
MAB_0528c|M.abscessus_ATCC_199 PAHIDDYNIEALSDDLLG-ILDDVGAGKATFVGHDWGAVVTWHTALAVPE
MLBr_02297|M.leprae_Br4923 PRGYDGWTLAGDTAGLIR----ALGHSSATLVGHAEGGLACWATALLHPR
* . :
Mvan_2384|M.vanbaalenii_PYR-1 RFARVVAANTTLPTG-----DQRPGEAF-------LAWQ-----RFSQET
Mflv_4004|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_6314|M.smegmatis_MC2_155 RFANIVVANTGLPTG-----DIPMPDIW-------WQFR-----NAIQKA
Mb2318|M.bovis_AF2122/97 RIARLVVANGFLPAA-----QGRTPLPF-------YVWR-----AFARYS
Rv2296|M.tuberculosis_H37Rv RIARLVVANGFLPAA-----QGRTPLPF-------YVWR-----AFARYS
MAV_2130|M.avium_104 RVGRLVVANGFLPTA-----QRRTPPAF-------YAWR-----AFARYS
MMAR_3472|M.marinum_M AIARLVVANGFLPTA-----RGRTPTAF-------HIWR-----AFARYS
MUL_3052|M.ulcerans_Agy99 RVRGIVLGNTWFWPA-----DTLTMKAF-------STVMSSLPMQYAILH
TH_1615|M.thermoresistible__bu RVQGIAHMESIVTPLTWADWEDPARSVF-------QGLRTP-EGERMVLE
MAB_0528c|M.abscessus_ATCC_199 RVSGVVGLSVPFTRRSQVPPTQAWNKLFGDNFFYILYFQEPGVADADLNR
MLBr_02297|M.leprae_Br4923 LVRAIALINSPHPAALRRSMLTHRNQGY-------ALLP-----TLLRYQ
Mvan_2384|M.vanbaalenii_PYR-1 PDFAVGHIVNGGCVSTLAPEVIAAYDAPFPDD-SYKAGARQFPTLVPTSP
Mflv_4004|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_6314|M.smegmatis_MC2_155 PKVDVGRFVASGCRRPLSDGVIAAYDAPFPDD-SYCAGPRAMPGLVPTSP
Mb2318|M.bovis_AF2122/97 PVLPAGRLVNFGTVHRVPAGVRAGYDAPFPDK-TYQAGARAFPRLVPTSP
Rv2296|M.tuberculosis_H37Rv PVLPAGRLVNFGTVHRVPAGVRAGYDAPFPDK-TYQAGARAFPRLVPTSP
MAV_2130|M.avium_104 PVLPAGRIVSVGTVRRVSSKVRAGYDAPFPDK-TYQAGARAFPQLVPTSP
MMAR_3472|M.marinum_M PVLPAGRLVAAGTVRKVPPAVRAGYDAPFPDK-SYQAGARAFPQLVPISP
MUL_3052|M.ulcerans_Agy99 HNFFVERLIPAGTTHRPSSAVMEHYRGVQPTA-SARVGVAEMPKQILAAR
TH_1615|M.thermoresistible__bu ENIFVEAMLPRGVHRTLSDEEMAHYREPFRDRGEGRRPTLTWPRNVPIDG
MAB_0528c|M.abscessus_ATCC_199 DPATTMRRMMAGMARIDGATMIAPGPAGFVERMPDPGELPEWLSQDELDH
MLBr_02297|M.leprae_Br4923 LPIWPERLLTRNNAAEIERLVRSRSSAKWQACEDFAVTIDHLRIAIQIPA
Mvan_2384|M.vanbaalenii_PYR-1 DDPAAPAN----------------RAAWEVLRTF-DRPFLCAFSDSDPIT
Mflv_4004|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_6314|M.smegmatis_MC2_155 DDPAAEAN----------------RAAWKVLRAS-KTPMLVAFSDGDPIT
Mb2318|M.bovis_AF2122/97 DDPAVPAN----------------RAAWEALGRW-DKPFLAIFGYRDPIL
Rv2296|M.tuberculosis_H37Rv DDPAVPAN----------------RAAWEALGRW-DKPFLAIFGYRDPIL
MAV_2130|M.avium_104 ADPAIPAN----------------RKAWEALGRW-EKPFLAIFGARDPIL
MMAR_3472|M.marinum_M DDPAVAAN----------------RAAWDALGRW-EKPFLAIFGERDPLL
MUL_3052|M.ulcerans_Agy99 PLLARLAS----------------EVPAQLG----TKPALLVWGMKDFAF
TH_1615|M.thermoresistible__bu EPADVAAI----------------VADYAVWLAEGDVPKLFVNAEPGAIV
MAB_0528c|M.abscessus_ATCC_199 YITEFARTGFTGGLNWYRNFDRNWALTERLAGANVVVPSLFIAGAADPVL
MLBr_02297|M.leprae_Br4923 AAHCALEYQRWAVRSQLRN---EGRKFMKSMSRPFSVPLLHVRGDADPYL
Mvan_2384|M.vanbaalenii_PYR-1 RGADAPLRKFIPGAQGRPEVTIAGGGHFLQEDKGPELAAVVVDFIAANPV
Mflv_4004|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_6314|M.smegmatis_MC2_155 GGMAPIFKKEMRGAHGIEHPVISGAGHFLQEDAGEDLASAVVEFLRDR--
Mb2318|M.bovis_AF2122/97 GQADGPLIKHIPGAAGQPHARIK-ASHFIQEDSGTELAERMLSWQQAT--
Rv2296|M.tuberculosis_H37Rv GQADGPLIKHIPGAAGQPHARIK-ASHFIQEDSGTELAERMLSWQQAT--
MAV_2130|M.avium_104 GHADSPLIKHIPGAAGQPHARIN-ASHFIQEDRGPELAERILSWQQALL-
MMAR_3472|M.marinum_M GRADRPLIKHIPGAAGQPHARIN-ANHFIQEDSGPELAERIISWQ-----
MUL_3052|M.ulcerans_Agy99 R-PEPMLARMRAVFADHILVELPEAKHLIQEDAPDEIAAAIIARFG----
TH_1615|M.thermoresistible__bu RGRVRDFVRSWP----NQTEITVAGRHYIQEDSPDEIGAAIADFVRKLRG
MAB_0528c|M.abscessus_ATCC_199 GFTDHAGSAKYR-TDNRGDLLIDGAGHWIQQERPLEVNAALLAFLREVVH
MLBr_02297|M.leprae_Br4923 LADSVDRTRRYAPHG--RYVSVSDAGHFSHEEAPEEINRYLTRFLKQVHG
Mvan_2384|M.vanbaalenii_PYR-1 ----
Mflv_4004|M.gilvum_PYR-GCK ----
MSMEG_6314|M.smegmatis_MC2_155 ----
Mb2318|M.bovis_AF2122/97 ----
Rv2296|M.tuberculosis_H37Rv ----
MAV_2130|M.avium_104 ----
MMAR_3472|M.marinum_M ----
MUL_3052|M.ulcerans_Agy99 ----
TH_1615|M.thermoresistible__bu ----
MAB_0528c|M.abscessus_ATCC_199 ----
MLBr_02297|M.leprae_Br4923 PRLS