For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDLYSNLVEAEQRLVALVSSIEADSYSSPTPCDRWDVRALLSHALASIDAFAAAVDGAPGPDMAQVFSGA DIVGDDPLGATQRITRRSQAAWSTVRDLNAELSTFIGVMPAGQALAIITFSTVVHGWDLAVATGQAGELP EHLAEAAQQVAAELVPVLRPRGLFAHDVDLAGEATPTQRLVALTGRKPR
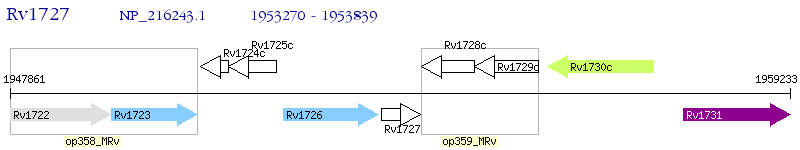
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv1727 | - | - | 100% (189) | hypothetical protein Rv1727 |
| M. tuberculosis H37Rv | Rv3773c | - | 6e-12 | 30.00% (180) | hypothetical protein Rv3773c |
| M. tuberculosis H37Rv | Rv0738 | - | 6e-12 | 26.49% (185) | hypothetical protein Rv0738 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1756 | - | 1e-104 | 100.00% (189) | hypothetical protein Mb1756 |
| M. gilvum PYR-GCK | Mflv_1118 | - | 4e-08 | 28.98% (176) | hypothetical protein Mflv_1118 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2430c | - | 5e-16 | 33.87% (186) | hypothetical protein MAB_2430c |
| M. marinum M | MMAR_1076 | - | 7e-12 | 24.87% (189) | hypothetical protein MMAR_1076 |
| M. avium 104 | MAV_0248 | - | 8e-17 | 35.50% (169) | hypothetical protein MAV_0248 |
| M. smegmatis MC2 155 | MSMEG_6832 | - | 4e-14 | 31.58% (190) | hypothetical protein MSMEG_6832 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0834 | - | 2e-12 | 26.70% (191) | hypothetical protein MUL_0834 |
| M. vanbaalenii PYR-1 | Mvan_2932 | - | 1e-12 | 29.79% (188) | hypothetical protein Mvan_2932 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1118|M.gilvum_PYR-GCK -------------------------MIDMTAACAGTAGLLDRVRDDQLSG
MAB_2430c|M.abscessus_ATCC_199 ----------------------MEIPFSLRPAASAVAALLPGISDSALDN
Rv1727|M.tuberculosis_H37Rv ----------------------MDLYSNLVEAEQRLVALVSSIEADSYSS
Mb1756|M.bovis_AF2122/97 ----------------------MDLYSNLVEAEQRLVALVSSIEADSYSS
MAV_0248|M.avium_104 ------MANMAPDLRPGPDSPPTDELDGAEAALRVMQQVLHPIAADDMPR
MSMEG_6832|M.smegmatis_MC2_155 -------MTYAGARASTPHWADMDELSSAQSALSALRPVLSAITPDDCGL
MMAR_1076|M.marinum_M ----------------------MDPLTAHQRAQDAFGSVLANVSADQLGA
MUL_0834|M.ulcerans_Agy99 ----------------------MDPLTAHQRAQDAFGSVLANVSADQLGA
Mvan_2932|M.vanbaalenii_PYR-1 MTGLGRHNGAMIAPTSSTTDHLKDPRPILDRAIATGGVVIAGVRPEQLTD
* :: : .
Mflv_1118|M.gilvum_PYR-GCK ATPCSGMDLGALVAHIGGLALAF---DAAARKEFGPLTDTPPEMGDGVED
MAB_2430c|M.abscessus_ATCC_199 PTPCTELTVRALVNHVMGLSLAFRYAAAPAEAAAAGFVSSGPSFTEDLDP
Rv1727|M.tuberculosis_H37Rv PTPCDRWDVRALLSHALASIDAFA---AAVDGAPGPDMAQVFSGADIVGD
Mb1756|M.bovis_AF2122/97 PTPCDRWDVRALLSHALASIDAFA---AAVDGAPGPDMAQVFSGADIVGD
MAV_0248|M.avium_104 PTPCAQFDVTRLTDHLLKSLEALG---GMAG-------ADFP--DHADSA
MSMEG_6832|M.smegmatis_MC2_155 PTPCAGFDVAALGEHLVGTVRMVG---AAAD-------AGFVRFEVVSHD
MMAR_1076|M.marinum_M ATPCSEWTVSDLIEHVIGGNEHVG------------IWSGGADRPAARPD
MUL_0834|M.ulcerans_Agy99 ATPCSEWTVSDLIEHVIGGNEHVG------------IWSGGADRPAARPD
Mvan_2932|M.vanbaalenii_PYR-1 PTPCTDMDVRALLAHLIGVLARVAALGNGEN-------PFAVTETSVADD
.*** : * * .
Mflv_1118|M.gilvum_PYR-GCK GWRESYPSRLASLASAWREPAAWEGMSRAGGVDFPADVGGLIALTEVVVH
MAB_2430c|M.abscessus_ATCC_199 QWRTSLPRRLADLVEVWERPVSWEGTSTIAGNVMPNHQVAMVALDELVLH
Rv1727|M.tuberculosis_H37Rv DPLGATQRITRRSQAAWSTVRDLNAELSTFIGVMPAGQALAIITFSTVVH
Mb1756|M.bovis_AF2122/97 DPLGATQRITRRSQAAWSTVRDLNAELSTFIGVMPAGQALAIITFSTVVH
MAV_0248|M.avium_104 DSVERRVIAVARPALDAWRQRGLDGTVSFGGGEMPARNACAILALELLVH
MSMEG_6832|M.smegmatis_MC2_155 DDIEARVAASTEAVIAVWQRRGTAGEVVFAGRVMPARLALGVLSVELVVH
MMAR_1076|M.marinum_M DMVAAHRATAAAAQQVFAAPDGMATVFKLPFGEIPGQVFIGMRTSDVLTH
MUL_0834|M.ulcerans_Agy99 DMVAAHRATAAAAQQVFAAPDGMATVFKLPFGEIPGQVFIGMRTSDVLTH
Mvan_2932|M.vanbaalenii_PYR-1 RWLDAWRQSGRRAADAWSDDAVLERPMALPWIQGSGAEVLASYFSELTVH
. . *
Mflv_1118|M.gilvum_PYR-GCK GWDVAVSAGLPYEVDGETLAAVHGHLAAFT-ADGP--------------V
MAB_2430c|M.abscessus_ATCC_199 GWDLAVATGQPFELPEGTDPGLFGFITDLA-SDGG--------------I
Rv1727|M.tuberculosis_H37Rv GWDLAVATGQAGELPEHLAEAAQQVAAELV-PVLR--------------P
Mb1756|M.bovis_AF2122/97 GWDLAVATGQAGELPEHLAEAAQQVAAELV-PVLR--------------P
MAV_0248|M.avium_104 AWDYARAVGRDARAPEPLAEYVLGLAHRVIRPEVR--------------G
MSMEG_6832|M.smegmatis_MC2_155 GWDFAQALHRPLGITAAHADFVLAMARHIITPASR--------------E
MMAR_1076|M.marinum_M AWDLAVATGQPSDLDPDLATQQLAAVRAFVGPQFR--------------G
MUL_0834|M.ulcerans_Agy99 AWDLAVATGQPSDLDPDLATQQLAAVRVFVGPQFR--------------G
Mvan_2932|M.vanbaalenii_PYR-1 TWDLATATRQQPDWDDTVVAAAFDAARQILPAENRRALYEEISAARGLDE
** * : . .
Mflv_1118|M.gilvum_PYR-GCK EG-LFDAPVPVPDDAPLLDRVIAMTGRDPSWSAA-----
MAB_2430c|M.abscessus_ATCC_199 PG-LFGPRVSVAPDAPRFDRMLAMSGRDPGWQPEVM---
Rv1727|M.tuberculosis_H37Rv RG-LFAHDVDLAGEATPTQRLVALTGRKPR---------
Mb1756|M.bovis_AF2122/97 RG-LFAHDVDLAGEATPTQRLVALTGRKPR---------
MAV_0248|M.avium_104 QA-GFDDPVEVPADADALTKLVAFTGRNPAR--------
MSMEG_6832|M.smegmatis_MC2_155 TA-GFDDPVSVAADAAALDRLVAYTGRTPRRWPSNMKSR
MMAR_1076|M.marinum_M PGKPFGQEQPCSAELSPADQLAAFLGRKVQ---------
MUL_0834|M.ulcerans_Agy99 PGKPFGQEQPCSAELSPADQLAAFLGRKVQ---------
Mvan_2932|M.vanbaalenii_PYR-1 VAVPFAEAVPISDDAPAIDRLVAWNGRDPLRRF------
. * . : :: * **