For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDPLMAHQRAQDAFAALLANVRADQLGGPTPCSEWTINDLIEHVVGGNEQVGRWAASPIEPPARPDGLVA AHQAAAAVAHEIFAAPGGMSATFKLPLGEVPGQVFIGLRTTDVLTHAWDLAAATGQSTDLDPELAVERLA AARALVGPQFRGPGKPFADEKPCPRERPPADQLAAFLGRTVR
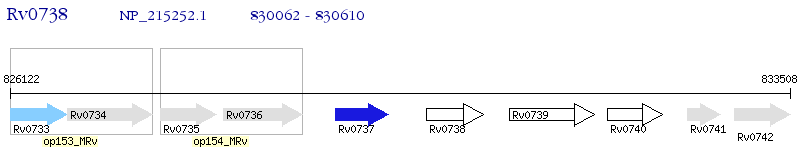
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0738 | - | - | 100% (182) | hypothetical protein Rv0738 |
| M. tuberculosis H37Rv | Rv0576 | - | 5e-12 | 29.28% (181) | ArsR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv1727 | - | 6e-12 | 26.49% (185) | hypothetical protein Rv1727 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0759 | - | 1e-104 | 100.00% (182) | hypothetical protein Mb0759 |
| M. gilvum PYR-GCK | Mflv_1118 | - | 8e-09 | 30.73% (179) | hypothetical protein Mflv_1118 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3926 | - | 5e-24 | 38.51% (174) | hypothetical protein MAB_3926 |
| M. marinum M | MMAR_1076 | - | 2e-75 | 70.33% (182) | hypothetical protein MMAR_1076 |
| M. avium 104 | MAV_0248 | - | 4e-13 | 27.22% (180) | hypothetical protein MAV_0248 |
| M. smegmatis MC2 155 | MSMEG_6832 | - | 5e-10 | 27.42% (186) | hypothetical protein MSMEG_6832 |
| M. thermoresistible (build 8) | TH_0767 | - | 5e-05 | 28.38% (148) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0834 | - | 4e-75 | 69.78% (182) | hypothetical protein MUL_0834 |
| M. vanbaalenii PYR-1 | Mvan_1558 | - | 4e-12 | 29.17% (168) | hypothetical protein Mvan_1558 |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0738|M.tuberculosis_H37Rv --------------------MDP-------LMAHQ---------------
Mb0759|M.bovis_AF2122/97 --------------------MDP-------LMAHQ---------------
MMAR_1076|M.marinum_M --------------------MDP-------LTAHQ---------------
MUL_0834|M.ulcerans_Agy99 --------------------MDP-------LTAHQ---------------
MAB_3926|M.abscessus_ATCC_1997 MSASEEGQALMSASEEGRALMSASEEGRALMSASEEGRAATSGASAEDFA
MAV_0248|M.avium_104 -----------MANMAPDLRPGPDSPPTDELDGAE---------------
MSMEG_6832|M.smegmatis_MC2_155 ------------MTYAGARASTPHWADMDELSSAQ---------------
Mvan_1558|M.vanbaalenii_PYR-1 ---------------------------MPMIRDQHR--------------
Mflv_1118|M.gilvum_PYR-GCK ------------------------------MIDMT---------------
TH_0767|M.thermoresistible__bu ------------------------------VDFRQ---------------
:
Rv0738|M.tuberculosis_H37Rv RAQDAFAALLANVRADQLGGPTP-CSEWTINDLIEHVVGGN-EQVGRWAA
Mb0759|M.bovis_AF2122/97 RAQDAFAALLANVRADQLGGPTP-CSEWTINDLIEHVVGGN-EQVGRWAA
MMAR_1076|M.marinum_M RAQDAFGSVLANVSADQLGAATP-CSEWTVSDLIEHVIGGN-EHVGIWSG
MUL_0834|M.ulcerans_Agy99 RAQDAFGSVLANVSADQLGAATP-CSEWTVSDLIEHVIGGN-EHVGIWSG
MAB_3926|M.abscessus_ATCC_1997 RASDAIEALLAAVRPDQWDAATP-CEEWNLRQLADHLVEVNYSLAGRFGG
MAV_0248|M.avium_104 AALRVMQQVLHPIAADDMPRPTP-CAQFDVTRLTDHLLKSLEALGGMAGA
MSMEG_6832|M.smegmatis_MC2_155 SALSALRPVLSAITPDDCGLPTP-CAGFDVAALGEHLVGTVRMVGAAADA
Mvan_1558|M.vanbaalenii_PYR-1 TAVQASIDVVAAVRPEHLTLDTP-CAGWNLADLLTHMTAQHHGFAAAARG
Mflv_1118|M.gilvum_PYR-GCK AACAGTAGLLDRVRDDQLSGATP-CSGMDLGALVAHIGGLALAFDAAARK
TH_0767|M.thermoresistible__bu VLLDETRAFGELIRSGDPQTPVPTCGEWTLRQLFRHVGRGHRWAAQIVSD
. : . .* * : * *:
Rv0738|M.tuberculosis_H37Rv SPIE-----------PPARPDGLVAAHQAAAAVAHEIFAAPGGMSATFKL
Mb0759|M.bovis_AF2122/97 SPIE-----------PPARPDGLVAAHQAAAAVAHEIFAAPGGMSATFKL
MMAR_1076|M.marinum_M GADR-----------PAARPDDMVAAHRATAAAAQQVFAAPDGMATVFKL
MUL_0834|M.ulcerans_Agy99 GADR-----------PAARPDDMVAAHRATAAAAQQVFAAPDGMATVFKL
MAB_3926|M.abscessus_ATCC_1997 LSSG-----------TAADP---VAAYRLSAQALRDALALPGVLDQTYPG
MAV_0248|M.avium_104 DFP------------DHADSADSVERRVIAVARPALDAWRQRGLDGTVSF
MSMEG_6832|M.smegmatis_MC2_155 GFVRF----------EVVSHDDDIEARVAASTEAVIAVWQRRGTAGEVVF
Mvan_1558|M.vanbaalenii_PYR-1 GGADLSAWRTE--PFADAVAADPAGTYAAAAHAVLAAFAADGIDEAPFAL
Mflv_1118|M.gilvum_PYR-GCK EFGPLT-------DTPPEMGDGVEDGWRESYPSRLASLASAWREPAAWEG
TH_0767|M.thermoresistible__bu RLSEPLDPRAVRDGKPPEDIEAALRWLQGGAQLVLDAVEAVGAGARVWTF
Rv0738|M.tuberculosis_H37Rv PLG----EVPGQVFIGLRTTDVLTHAWDLAAATGQSTDLDPELAVERLAA
Mb0759|M.bovis_AF2122/97 PLG----EVPGQVFIGLRTTDVLTHAWDLAAATGQSTDLDPELAVERLAA
MMAR_1076|M.marinum_M PFG----EIPGQVFIGMRTSDVLTHAWDLAVATGQPSDLDPDLATQQLAA
MUL_0834|M.ulcerans_Agy99 PFG----EIPGQVFIGMRTSDVLTHAWDLAVATGQPSDLDPDLATQQLAA
MAB_3926|M.abscessus_ATCC_1997 PFA----HTTGANQLQVRMADLLTHGWDLARATGASADLPVDLTENALGF
MAV_0248|M.avium_104 GGG----EMPARNACAILALELLVHAWDYARAVGRDARAPEPLAEYVLGL
MSMEG_6832|M.smegmatis_MC2_155 AGR----VMPARLALGVLSVELVVHGWDFAQALHRPLGITAAHADFVLAM
Mvan_1558|M.vanbaalenii_PYR-1 PEFGPGVSVPGSTAMGFHFVDYVVHGWDVAATLRTPYTLPPDVLAAALPI
Mflv_1118|M.gilvum_PYR-GCK MSRAGGVDFPADVGGLIALTEVVVHGWDVAVSAGLPYEVDGETLAAVHG-
TH_0767|M.thermoresistible__bu LGP-----RPAGWWVRRRVHETTVHRADAALALGAEYTLPAELAADAISE
.. : .* * * :
Rv0738|M.tuberculosis_H37Rv ARALVGPQFRGPGKP--------------------------------FAD
Mb0759|M.bovis_AF2122/97 ARALVGPQFRGPGKP--------------------------------FAD
MMAR_1076|M.marinum_M VRAFVGPQFRGPGKP--------------------------------FGQ
MUL_0834|M.ulcerans_Agy99 VRVFVGPQFRGPGKP--------------------------------FGQ
MAB_3926|M.abscessus_ATCC_1997 VQKLAG-AFARSGK---------------------------------FGA
MAV_0248|M.avium_104 AHRVIRPEVRGQAG---------------------------------FDD
MSMEG_6832|M.smegmatis_MC2_155 ARHIITPASRETAG---------------------------------FDD
Mvan_1558|M.vanbaalenii_PYR-1 VMTVPDGEFRDADNSP-------------------------------FAR
Mflv_1118|M.gilvum_PYR-GCK --HLAAFTADGPVEGL-------------------------------FDA
TH_0767|M.thermoresistible__bu WIELMAVQARRTQLPLERGRSLHLHATDAGLGAVGEWLITSDEEGLDWTH
. :
Rv0738|M.tuberculosis_H37Rv EK---PCPRERPPADQLAAFLGRTVR-----------------------
Mb0759|M.bovis_AF2122/97 EK---PCPRERPPADQLAAFLGRTVR-----------------------
MMAR_1076|M.marinum_M EQ---PCSAELSPADQLAAFLGRKVQ-----------------------
MUL_0834|M.ulcerans_Agy99 EQ---PCSAELSPADQLAAFLGRKVQ-----------------------
MAB_3926|M.abscessus_ATCC_1997 PQ---PVAEDAPALDRLAAMTGRVV------------------------
MAV_0248|M.avium_104 PV---EVPADADALTKLVAFTGRNPAR----------------------
MSMEG_6832|M.smegmatis_MC2_155 PV---SVAADAAALDRLVAYTGRTPRRWPSNMKSR--------------
Mvan_1558|M.vanbaalenii_PYR-1 AL---SLDDTDDDLARILRHLGRNPEYC---------------------
Mflv_1118|M.gilvum_PYR-GCK PV---PVPDDAPLLDRVIAMTGRDPSWSAA-------------------
TH_0767|M.thermoresistible__bu EHGKGDVALRGPATDLLLALSRRRTVDDLGVELHGDIAVWERWLANTSL
: *