For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
EIPFSLRPAASAVAALLPGISDSALDNPTPCTELTVRALVNHVMGLSLAFRYAAAPAEAAAAGFVSSGPS FTEDLDPQWRTSLPRRLADLVEVWERPVSWEGTSTIAGNVMPNHQVAMVALDELVLHGWDLAVATGQPFE LPEGTDPGLFGFITDLASDGGIPGLFGPRVSVAPDAPRFDRMLAMSGRDPGWQPEVM*
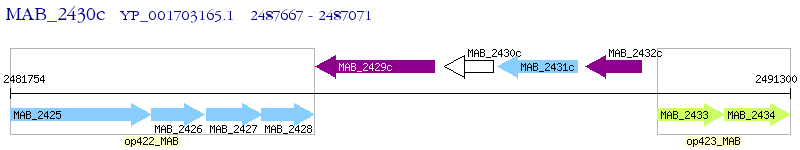
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_2430c | - | - | 100% (198) | hypothetical protein MAB_2430c |
| M. abscessus ATCC 19977 | MAB_3926 | - | 4e-14 | 30.00% (180) | hypothetical protein MAB_3926 |
| M. abscessus ATCC 19977 | MAB_4446 | - | 7e-13 | 26.74% (187) | hypothetical protein MAB_4446 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1756 | - | 5e-16 | 33.87% (186) | hypothetical protein Mb1756 |
| M. gilvum PYR-GCK | Mflv_1118 | - | 4e-34 | 38.59% (184) | hypothetical protein Mflv_1118 |
| M. tuberculosis H37Rv | Rv1727 | - | 5e-16 | 33.87% (186) | hypothetical protein Rv1727 |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_5328 | - | 1e-08 | 25.53% (188) | hypothetical protein MMAR_5328 |
| M. avium 104 | MAV_0248 | - | 1e-07 | 24.61% (191) | hypothetical protein MAV_0248 |
| M. smegmatis MC2 155 | MSMEG_6832 | - | 4e-13 | 34.33% (201) | hypothetical protein MSMEG_6832 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4409 | - | 1e-08 | 26.06% (188) | hypothetical protein MUL_4409 |
| M. vanbaalenii PYR-1 | Mvan_5694 | - | 2e-30 | 36.41% (184) | hypothetical protein Mvan_5694 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1118|M.gilvum_PYR-GCK -------------------MIDMTAACAGTAGLLDRVRDDQLSGATPCSG
Mvan_5694|M.vanbaalenii_PYR-1 -------------------MIDMTEACTTTAELLAQVSDDQLTAATPCRD
MAB_2430c|M.abscessus_ATCC_199 ----------------MEIPFSLRPAASAVAALLPGISDSALDNPTPCTE
Mb1756|M.bovis_AF2122/97 ----------------MDLYSNLVEAEQRLVALVSSIEADSYSSPTPCDR
Rv1727|M.tuberculosis_H37Rv ----------------MDLYSNLVEAEQRLVALVSSIEADSYSSPTPCDR
MMAR_5328|M.marinum_M ---MASDLRPGPDSPPTDELASAERALAVLQQVLHPIGDGDLFRPTPCPE
MUL_4409|M.ulcerans_Agy99 ---MASDLRPGPDSPPTDELASAERALAVLQQVLHPIGDGDLFRPTPCPE
MAV_0248|M.avium_104 MANMAPDLRPGPDSPPTDELDGAEAALRVMQQVLHPIAADDMPRPTPCAQ
MSMEG_6832|M.smegmatis_MC2_155 -MTYAGARASTPHWADMDELSSAQSALSALRPVLSAITPDDCGLPTPCAG
. * :: : . .***
Mflv_1118|M.gilvum_PYR-GCK MDLGALVAHIGGLALAF---DAAARKEFGPLTDTPPEMGDGVEDGWRESY
Mvan_5694|M.vanbaalenii_PYR-1 MDLGALIAHVGGLALAF---EAAARKDFGPLTDTPPTMEGGVDDGWRQAY
MAB_2430c|M.abscessus_ATCC_199 LTVRALVNHVMGLSLAFRYAAAPAEAAAAGFVSSGPSFTEDLDPQWRTSL
Mb1756|M.bovis_AF2122/97 WDVRALLSHALASIDAF---AAAVDGAPGPDMAQVFSGADIVGDDPLGAT
Rv1727|M.tuberculosis_H37Rv WDVRALLSHALASIDAF---AAAVDGAPGPDMAQVFSGADIVGDDPLGAT
MMAR_5328|M.marinum_M YDVSGLTGHLLNSIVVLG---GMAGAEFT---------MRDHTSSAEGQV
MUL_4409|M.ulcerans_Agy99 YDVSGLTGHLLNSIVVLD---GMAGAEFT---------MRDHTSTPEGQV
MAV_0248|M.avium_104 FDVTRLTDHLLKSLEALG---GMAGADFPD--------HADSADSVERRV
MSMEG_6832|M.smegmatis_MC2_155 FDVAALGEHLVGTVRMVG---AAADAGFVRFE------VVSHDDDIEARV
: * * . . .
Mflv_1118|M.gilvum_PYR-GCK PSRLASLASAWREPAAWEGMSRAGGVDFPADVGGLIALTEVVVHGWDVAV
Mvan_5694|M.vanbaalenii_PYR-1 PSRLAALAQAWRDPAAWQGMSRAGGVDFPAELGGMIALTEVVVHGWDVAV
MAB_2430c|M.abscessus_ATCC_199 PRRLADLVEVWERPVSWEGTSTIAGNVMPNHQVAMVALDELVLHGWDLAV
Mb1756|M.bovis_AF2122/97 QRITRRSQAAWSTVRDLNAELSTFIGVMPAGQALAIITFSTVVHGWDLAV
Rv1727|M.tuberculosis_H37Rv QRITRRSQAAWSTVRDLNAELSTFIGVMPAGQALAIITFSTVVHGWDLAV
MMAR_5328|M.marinum_M VSAARPALDAWHR-RGMEGDVPLGPASMPARVAVSVFSIEFLVHAWDYAT
MUL_4409|M.ulcerans_Agy99 VSAARPALDAWHR-RGMDGDVPLGPASMPARVAVSVFSIEFLVHAWDYAT
MAV_0248|M.avium_104 IAVARPALDAWRQ-RGLDGTVSFGGGEMPARNACAILALELLVHAWDYAR
MSMEG_6832|M.smegmatis_MC2_155 AASTEAVIAVWQR-RGTAGEVVFAGRVMPARLALGVLSVELVVHGWDFAQ
.* . :* : . ::*.** *
Mflv_1118|M.gilvum_PYR-GCK SAGLPYEVDGETLAAVHGHLAAFTA-DGPVEGLFDAPVPVPDDAPLLDRV
Mvan_5694|M.vanbaalenii_PYR-1 SAGLRYDVPVDVLNVVLPHVAAFAE-QGPVEGLFAAPVPVPGDAPVLHRV
MAB_2430c|M.abscessus_ATCC_199 ATGQPFELPEGTDPGLFGFITDLAS-DGGIPGLFGPRVSVAPDAPRFDRM
Mb1756|M.bovis_AF2122/97 ATGQAGELPEHLAEAAQQVAAELVP-VLRPRGLFAHDVDLAGEATPTQRL
Rv1727|M.tuberculosis_H37Rv ATGQAGELPEHLAEAAQQVAAELVP-VLRPRGLFAHDVDLAGEATPTQRL
MMAR_5328|M.marinum_M AVGHDIHVPDSLADYVLGLAQNLIQPHERGAAGFDDPVQVGDDVSGLHRL
MUL_4409|M.ulcerans_Agy99 AVGHDIHVPDSLADYVLGLAQNLIQPHERGAAGFDDPVQVGDDVRGLHRL
MAV_0248|M.avium_104 AVGRDARAPEPLAEYVLGLAHRVIRPEVRGQAGFDDPVEVPADADALTKL
MSMEG_6832|M.smegmatis_MC2_155 ALHRPLGITAAHADFVLAMARHIITPASRETAGFDDPVSVAADAAALDRL
: . . * * : :. ::
Mflv_1118|M.gilvum_PYR-GCK IAMTGRDPSWSAA-----
Mvan_5694|M.vanbaalenii_PYR-1 LGLTGRDPSWPAG-----
MAB_2430c|M.abscessus_ATCC_199 LAMSGRDPGWQPEVM---
Mb1756|M.bovis_AF2122/97 VALTGRKPR---------
Rv1727|M.tuberculosis_H37Rv VALTGRKPR---------
MMAR_5328|M.marinum_M VAFTGRNPIR--------
MUL_4409|M.ulcerans_Agy99 VAFTGRNPIR--------
MAV_0248|M.avium_104 VAFTGRNPAR--------
MSMEG_6832|M.smegmatis_MC2_155 VAYTGRTPRRWPSNMKSR
:. :** *