For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TYAGARASTPHWADMDELSSAQSALSALRPVLSAITPDDCGLPTPCAGFDVAALGEHLVGTVRMVGAAAD AGFVRFEVVSHDDDIEARVAASTEAVIAVWQRRGTAGEVVFAGRVMPARLALGVLSVELVVHGWDFAQAL HRPLGITAAHADFVLAMARHIITPASRETAGFDDPVSVAADAAALDRLVAYTGRTPRRWPSNMKSR*
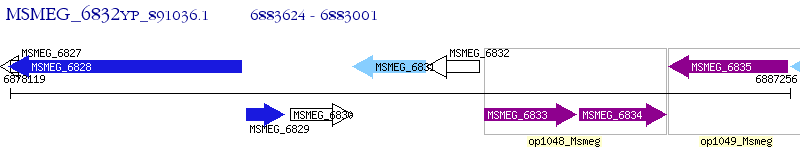
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6832 | - | - | 100% (207) | hypothetical protein MSMEG_6832 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0591 | - | 1e-34 | 39.69% (194) | ArsR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_1118 | - | 1e-10 | 30.85% (188) | hypothetical protein Mflv_1118 |
| M. tuberculosis H37Rv | Rv0576 | - | 1e-34 | 40.00% (200) | ArsR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4446 | - | 4e-23 | 33.51% (188) | hypothetical protein MAB_4446 |
| M. marinum M | MMAR_5328 | - | 5e-36 | 40.98% (183) | hypothetical protein MMAR_5328 |
| M. avium 104 | MAV_0248 | - | 7e-38 | 44.26% (183) | hypothetical protein MAV_0248 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_0693 | - | 1e-35 | 40.66% (182) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1558 | - | 3e-09 | 28.72% (195) | hypothetical protein Mvan_1558 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5328|M.marinum_M --------------------------------------------------
MAV_0248|M.avium_104 --------------------------------------------------
Mb0591|M.bovis_AF2122/97 MLEVAAEPTRRRLLQLLAPGERTVTQLASQFTVTRSAISQHLGMLAEAGL
Rv0576|M.tuberculosis_H37Rv MLEVAAEPTRRRLLQLLAPGERTVTQLASQFTVTRSAISQHLGMLAEAGL
MUL_0693|M.ulcerans_Agy99 --------------------------MNSPESLDSGYEPAPPGRPAET--
MAB_4446|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6832|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1558|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5328|M.marinum_M --------------------------------------------------
MAV_0248|M.avium_104 --------------------------------------------------
Mb0591|M.bovis_AF2122/97 VTARKQGRERYYRLDERGVLRLRALMESFWSDELDRLVADAAHYPPSQGD
Rv0576|M.tuberculosis_H37Rv VTARKQGRERYYRLDERGVLRLRALMESFWSDELDRLVADAAHYPPSQGD
MUL_0693|M.ulcerans_Agy99 ----------------------------------------------VSGE
MAB_4446|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6832|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1558|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5328|M.marinum_M --------------------------------------------------
MAV_0248|M.avium_104 --------------------------------------------------
Mb0591|M.bovis_AF2122/97 CAMPFEKAVVVPLDPTSTFALITQPDRLRRWMAVAARIELRTGGAYRWTV
Rv0576|M.tuberculosis_H37Rv CAMPFEKAVVVPLDPTSTFALITQPDRLRRWMAVAARIELRTGGAYRWTV
MUL_0693|M.ulcerans_Agy99 RAMPLENAVVLPVDPAQSFALFTRPEGLRRWMAITARIEPYDAGVYRWTV
MAB_4446|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6832|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1558|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5328|M.marinum_M --------------------------------------------------
MAV_0248|M.avium_104 --------------------------------------------------
Mb0591|M.bovis_AF2122/97 TPGHSAAGTVIDVDPGKRVVFTWGWEDHGDPPPGGSTVTITLTPVDGGTE
Rv0576|M.tuberculosis_H37Rv TPGHSAAGTVIDVDPGKRVVFTWGWEDHGDPPPGGSTVTITLTPVDGGTE
MUL_0693|M.ulcerans_Agy99 TPGHTAVGAVVEADPGQRLVLSWGWEGHGDPAPGESTVTVTLAPAPDGTE
MAB_4446|M.abscessus_ATCC_1997 --------------------------------------------------
MSMEG_6832|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1558|M.vanbaalenii_PYR-1 --------------------------------------------------
MMAR_5328|M.marinum_M -----------------------------MASDLRPGPD-----SPPTDE
MAV_0248|M.avium_104 --------------------------MANMAPDLRPGPD-----SPPTDE
Mb0591|M.bovis_AF2122/97 VRLVHDGLTAQQAARHAKGWNHFLDRLVVAGQHGDAGPDEWAAAPDPLDE
Rv0576|M.tuberculosis_H37Rv VRLVHDGLTAQQAARHAKGWNHFLDRLVVAGQRGDAGPDEWAAAPDPLDE
MUL_0693|M.ulcerans_Agy99 IRLVHEGLTEQQAARHAEGWSHYLDRLVAAALQGDAGPDEWGAVPGPLDE
MAB_4446|M.abscessus_ATCC_1997 -------------------------------------------MTTALNP
MSMEG_6832|M.smegmatis_MC2_155 ---------------------------MTYAGARASTPH-----WADMDE
Mflv_1118|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1558|M.vanbaalenii_PYR-1 ----------------------------------------------MPMI
MMAR_5328|M.marinum_M LASAERALAVLQQVLHPIGDGDLFRPTPCPEYDVSGLTGHLLNSIVVLGG
MAV_0248|M.avium_104 LDGAEAALRVMQQVLHPIAADDMPRPTPCAQFDVTRLTDHLLKSLEALGG
Mb0591|M.bovis_AF2122/97 LSCAEATLAVLQHVLRGIGASDLTRQTPCTEYDVSQLADHLLRSLAIIGA
Rv0576|M.tuberculosis_H37Rv LSCAEATLAVLQHVLRGIGASDLTRQTPCTEYDVSQLADHLLRSLAIIGA
MUL_0693|M.ulcerans_Agy99 LSCAEATLAVVQHVLRALEPADLARQTPCSQYDVVQLADHLMRSLTIIGG
MAB_4446|M.abscessus_ATCC_1997 LETVANARAALHEVVSRLTEADNDKQTPNAKFTVAQLTDHLQNSIKLLGG
MSMEG_6832|M.smegmatis_MC2_155 LSSAQSALSALRPVLSAITPDDCGLPTPCAGFDVAALGEHLVGTVRMVGA
Mflv_1118|M.gilvum_PYR-GCK MIDMTAACAGTAGLLDRVRDDQLSGATPCSGMDLGALVAHIGGLALAFDA
Mvan_1558|M.vanbaalenii_PYR-1 RDQHRTAVQASIDVVAAVRPEHLTLDTPCAGWNLADLLTHMTAQHHGFAA
: :: : . ** . : * *: . .
MMAR_5328|M.marinum_M MAG---AEFT---MRDHTSSAEGQVVS--------AARPALDAWHR----
MAV_0248|M.avium_104 MAG---ADFPD--HADSADSVERRVIA--------VARPALDAWRQ----
Mb0591|M.bovis_AF2122/97 AAG---AQLA---PRDVDAPLETQVAD--------AAQAVMEAWRR----
Rv0576|M.tuberculosis_H37Rv AAG---AQLA---PRDVDAPLETQVAD--------AAQAVMEAWRR----
MUL_0693|M.ulcerans_Agy99 AAG---AQLP---PRDLDAPLETQVAD--------AGQAVLEAWRR----
MAB_4446|M.abscessus_ATCC_1997 AAG---VDIA----LTTEGSVADRLLP--------QSQAVVDAWQR----
MSMEG_6832|M.smegmatis_MC2_155 AAD---AGFVRFEVVSHDDDIEARVAA--------STEAVIAVWQR----
Mflv_1118|M.gilvum_PYR-GCK AAR---KEFGP--LTDTPPEMGDGVEDGWRESYPSRLASLASAWREP---
Mvan_1558|M.vanbaalenii_PYR-1 AARGGGADLSAWRTEPFADAVAADPAG----TYAAAAHAVLAAFAADGID
* : . .:
MMAR_5328|M.marinum_M RGMEGDVPLG-PASMPARVAVSVFSIEFLVHAWDYATAVGHDIHVPDSLA
MAV_0248|M.avium_104 RGLDGTVSFG-GGEMPARNACAILALELLVHAWDYARAVGRDARAPEPLA
Mb0591|M.bovis_AF2122/97 RGLAGTVELN-SNQVPATVPVGILCLEFLVHAWDFAIATGSQVIASEPVS
Rv0576|M.tuberculosis_H37Rv RGLAGTVELN-SNQVPATVPVGILCLEFLVHAWDFAIATGSQVIASEPVS
MUL_0693|M.ulcerans_Agy99 RGVDGTVELN-ANQVPAAVPVGILCLELLVHAWDFAFATDRRVVVSDTVT
MAB_4446|M.abscessus_ATCC_1997 RGIDGTVTLP-IGEYPAEVAVRILGSEFLVHAWDYAVATGQEFEPMDALT
MSMEG_6832|M.smegmatis_MC2_155 RGTAGEVVFA-GRVMPARLALGVLSVELVVHGWDFAQALHRPLGITAAHA
Mflv_1118|M.gilvum_PYR-GCK AAWEGMSRAG-GVDFPADVGGLIALTEVVVHGWDVAVSAGLPYEVDGETL
Mvan_1558|M.vanbaalenii_PYR-1 EAPFALPEFGPGVSVPGSTAMGFHFVDYVVHGWDVAATLRTPYTLPPDVL
. . *. . : :**.** * :
MMAR_5328|M.marinum_M DYVLGLAQNLIQP--HERGAAGFDDPVQVGDDVSGLHRLVAFTGRNPIR-
MAV_0248|M.avium_104 EYVLGLAHRVIRP--EVRGQAGFDDPVEVPADADALTKLVAFTGRNPAR-
Mb0591|M.bovis_AF2122/97 EYVLAVAGKVITP--ATRNSAGFAAPAAVGSFAPVLDRLIAFTGRQPT--
Rv0576|M.tuberculosis_H37Rv EYVLAVAGKVITP--ATRNSAGFAAPAAVGSFAPVLDRLIAFTGRQPT--
MUL_0693|M.ulcerans_Agy99 EYVLTVAGKVITP--AARNSAGFAAPVAVGAFAPVLDRLIAFTGRRPVHP
MAB_4446|M.abscessus_ATCC_1997 DGVLESVRMIIQP--ERRDGDFFADEVPVPDDSPNLVKLIAFTGRNPGWS
MSMEG_6832|M.smegmatis_MC2_155 DFVLAMARHIITP--ASRETAGFDDPVSVAADAAALDRLVAYTGRTPRRW
Mflv_1118|M.gilvum_PYR-GCK AAVHGHLAAFTAD--GPVEG-LFDAPVPVPDDAPLLDRVIAMTGRDPSWS
Mvan_1558|M.vanbaalenii_PYR-1 AAALPIVMTVPDGEFRDADNSPFARALSLDDTDDDLARILRHLGRNPEYC
. . * : * ::: ** *
MMAR_5328|M.marinum_M -------------
MAV_0248|M.avium_104 -------------
Mb0591|M.bovis_AF2122/97 AGHVSAT------
Rv0576|M.tuberculosis_H37Rv AGHVSAT------
MUL_0693|M.ulcerans_Agy99 TGLTSPIRADPGS
MAB_4446|M.abscessus_ATCC_1997 PAS----------
MSMEG_6832|M.smegmatis_MC2_155 PSNMKSR------
Mflv_1118|M.gilvum_PYR-GCK AA-----------
Mvan_1558|M.vanbaalenii_PYR-1 -------------