For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTLKVKGEGLGAQVTGVDPKNLDDITTDEIRDIVYTNKLVVLKDVHPSPREFIKLGRIIGQIVPYYEPMY HHEDHPEIFVSSTEEGQGVPKTGAFWHIDYMFMPEPFAFSMVLPLAVPGHDRGTYFIDLARVWQSLPAAK RDPARGTVSTHDPRRHIKIRPSDVYRPIGEVWDEINRTTPPIKWPTVIRHPKTGQEILYICATGTTKIED KDGNPVDPEVLQELMAATGQLDPEYQSPFIHTQHYQVGDIILWDNRVLMHRAKHGSAAGTLTTYRLTMLD GLKTPGYAA
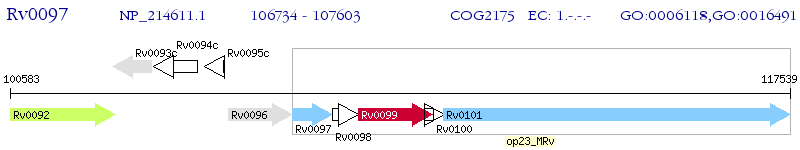
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. tuberculosis H37Rv | Rv0097 | - | - | 100% (289) | POSSIBLE OXIDOREDUCTASE |
| M. tuberculosis H37Rv | Rv3406 | - | 3e-09 | 23.97% (267) | dioxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0100 | - | 1e-174 | 100.00% (289) | oxidoreductase |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. leprae Br4923 | MLBr_01992 | - | 1e-136 | 76.39% (288) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0659 | - | 1e-138 | 76.82% (289) | putative dioxygenase |
| M. marinum M | MMAR_0260 | - | 1e-161 | 90.66% (289) | oxidoreductase |
| M. avium 104 | MAV_0235 | - | 2e-16 | 24.17% (302) | taurine catabolism dioxygenase TauD/TfdA |
| M. smegmatis MC2 155 | MSMEG_0181 | - | 9e-13 | 25.75% (268) | alpha-ketoglutarate-dependent taurine dioxygenase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4854 | - | 1e-159 | 89.62% (289) | oxidoreductase |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Rv0097|M.tuberculosis_H37Rv ------------------------------------------MTLKVKGE
Mb0100|M.bovis_AF2122/97 ------------------------------------------MTLKVKGE
MMAR_0260|M.marinum_M ------------------------------------------MTLNVKGE
MUL_4854|M.ulcerans_Agy99 ------------------------------------------MTLNVKGE
MAB_0659|M.abscessus_ATCC_1997 ------------------------------------------MTLNVKGE
MLBr_01992|M.leprae_Br4923 ------------------------------------------MTLDVKGD
MAV_0235|M.avium_104 ------------------------------------------MEIVASGP
MSMEG_0181|M.smegmatis_MC2_155 MTDTRHAPPPAPRPIPEYDVSAGPFSHLAAERDRLAALRWRHFDAEQVGT
: *
Rv0097|M.tuberculosis_H37Rv GLGAQVTGVDPKNLDDIT--TDEIRDIVYTNKLVVLKDVHPSPREFIKLG
Mb0100|M.bovis_AF2122/97 GLGAQVTGVDPKNLDDIT--TDEIRDIVYTNKLVVLKDVHPSPREFIKLG
MMAR_0260|M.marinum_M GLGAQITGVDPKNLDDIT--TDEIRDIVYANKLVILKDVNPSPEEFLKLG
MUL_4854|M.ulcerans_Agy99 GLGAQITGVDPKILDDIT--TDEIRDIVYANKLVILKDVNPSPEEFLKLG
MAB_0659|M.abscessus_ATCC_1997 GLGAQVTGIDPGNLDGIT--TEEIRELVYRHKLVVLKGVHPTPEQFIELG
MLBr_01992|M.leprae_Br4923 GLGAQVTGVDPKNLDSIS--TAEIRKIVYVNKLVVLKDVHPTPEEFIKLG
MAV_0235|M.avium_104 GFAAELRGVTIADVAASPDVYAQVRAAFEEHSVLVFRDQHVSDEAQLAFS
MSMEG_0181|M.smegmatis_MC2_155 TLGAQISGVDLTRPLAGA-VFDELRTALHEYKVLFFRDQPMSADAHAALA
:.*:: *: . ::* . .::.::. : :.
Rv0097|M.tuberculosis_H37Rv RIIGQIVPYYEPMYHHEDHPEIFVSSTEEGQGVP---------KTGAFWH
Mb0100|M.bovis_AF2122/97 RIIGQIVPYYEPMYHHEDHPEIFVSSTEEGQGVP---------KTGAFWH
MMAR_0260|M.marinum_M KIVGQIVPYYEPMYHHEDHPEIFVSSTEEGQGVP---------KTGAFWH
MUL_4854|M.ulcerans_Agy99 KIVGQIVPYYEPMYHHEDHPEIFVSSTVEGQGVP---------KTGAFWH
MAB_0659|M.abscessus_ATCC_1997 RLIGEIVPYYEPVYHHQDHPEIFVSSTEEGQGVP---------RTGAFWH
MLBr_01992|M.leprae_Br4923 RIIGEIVPYYEPIYRHKDYPEIFVSSTEEGQGVP---------NTGAFWH
MAV_0235|M.avium_104 RRFGPLGVTKVGAVGHGSHLVVLKTLDDDGNVVPADHRLALENKANQLWH
MSMEG_0181|M.smegmatis_MC2_155 RQFGPLEVH--PLLPANSAESALARFQKDANVSG----------YENAWH
: .* : . : :.: **
Rv0097|M.tuberculosis_H37Rv IDYMFMPEPFAFSMVLPLAVPGHDRGTYFIDLARVWQSLPAAKRDPARGT
Mb0100|M.bovis_AF2122/97 IDYMFMPEPFAFSMVLPLAVPGHDRGTYFIDLARVWQSLPAAKRDPARGT
MMAR_0260|M.marinum_M IDYMFMPEPFAFSMVLPLAVPGHDRGTYFIDLAKVWQSLPAAQQAPARGT
MUL_4854|M.ulcerans_Agy99 IDYMFMPEPFAFSMVLPLAVPGHDRGTYFIDLAKVWQSLPSAQQAPARGT
MAB_0659|M.abscessus_ATCC_1997 VDYMFMPKPFAFSMVLPLAVPGHDRGTYFIDLSKVWESLPAKTKDAALGT
MLBr_01992|M.leprae_Br4923 VDYIFMPEPFAFSMTLPLAMPGHDRGTHFIDLSQVWESLPNAMKDSVRGT
MAV_0235|M.avium_104 TDSSFKRVPALASVLSSRIVPGRGGETEYVSTRIAFERLDPGLRERVENS
MSMEG_0181|M.smegmatis_MC2_155 HDVTWRPEPSILAILHAIEVPPIGGDTLFADMYAAYDSLDDETKAEIDRL
* : * :: . :* . * : . .:: * :
Rv0097|M.tuberculosis_H37Rv VSTHDPRRHIKIRPSDVYRPIGEVWDEINRTTPPIKWPTVIRHPKTGQEI
Mb0100|M.bovis_AF2122/97 VSTHDPRRHIKIRPSDVYRPIGEVWDEINRTTPPIKWPTVIRHPKTGQEI
MMAR_0260|M.marinum_M LSTHDPRRHIKIRPSDVYRPIGEVWDEISRATPPIKWPTVIRHPKTGEEI
MUL_4854|M.ulcerans_Agy99 LSTHDPRRHIKIRPSDVYRPIGEVWDEISRATPPIKWPTVIRHPKTGEEI
MAB_0659|M.abscessus_ATCC_1997 FSTHTPRRYIKIRPSDVYRPIGEILAEIERTTPPQKWPTVIKHPKTGEEI
MLBr_01992|M.leprae_Br4923 YSNHSPRRYIKIRPSDVYRPVGEVLAEIEEVTPPQKWPTVIKHPKTGQEI
MAV_0235|M.avium_104 FAWHEYAYSRGKIAPDLARPE------ERAALPPQCWRLVWRNPVNGRKA
MSMEG_0181|M.smegmatis_MC2_155 DAAHDFTYYLRGLVPDE------KIAELQAAHPPVTHPVVCTHPITGRRH
: * .* . ** * :* .*..
Rv0097|M.tuberculosis_H37Rv LYICATGTTKIEDKDGNPVDPEVLQELMAATGQLDPEYQSPFIHTQHYQV
Mb0100|M.bovis_AF2122/97 LYICATGTTKIEDKDGNPVDPEVLQELMAATGQLDPEYQSPFIHTQHYQV
MMAR_0260|M.marinum_M LYICATGTTKIEDKDGNLVDPAVLAELLAATGQLDPEYNSPFIHTQHYEV
MUL_4854|M.ulcerans_Agy99 LYICATGTTKIEDKDGNLVDPAVLAELLAATGQLDPEYNSPFIHTQHYEV
MAB_0659|M.abscessus_ATCC_1997 LYICEAGTETIEDGAGRIQDAALLQQLLEASGQLDPDYASPFIHAQHYEV
MLBr_01992|M.leprae_Br4923 LYICEAATVSVEDKNGNLLDPMVLQELLTASGQLDPDCKSLLIHTQHYEV
MAV_0235|M.avium_104 LYLASHAYG-IEG-----MEPAAARELLAALTEAATAPGASYLHS--WRA
MSMEG_0181|M.smegmatis_MC2_155 LYVNRMFVSHIAG-----YPHDTGRALLDRLCRTADAPEHQVRFR--WAP
**: : . *: . . :
Rv0097|M.tuberculosis_H37Rv GDIILWDNRVLMHRAKHGSAAG-------TLTTYRLTMLDGLKTPGYAA-
Mb0100|M.bovis_AF2122/97 GDIILWDNRVLMHRAKHGSAAG-------TLTTYRLTMLDGLKTPGYAA-
MMAR_0260|M.marinum_M GDIILWDNRVLMHRAKHGTASG-------TLTTYRLTMLDGLETPGYPA-
MUL_4854|M.ulcerans_Agy99 GDIILWDNRVLMHRAKHGTASG-------TLTTYRLTMLDGLETPGYPA-
MAB_0659|M.abscessus_ATCC_1997 GDIVLWDNRSLVHRAKHGTASG-------TLTTYRLTMLDGLEAPGFAA-
MLBr_01992|M.leprae_Br4923 GDVVLWDNRALVHRAKHGTVSG-------TLITYRLTLLDGLKTPGYAV-
MAV_0235|M.avium_104 GDVVMWDNRATMHRGRPWPAHQPRCMVRSTIAATAADGLEAMYPPWHAVA
MSMEG_0181|M.smegmatis_MC2_155 DSVAIWDNRAVQHYAASG-----------YWPHTRIMERASVTGPVPCR-
..: :**** * . .: *
Rv0097|M.tuberculosis_H37Rv -
Mb0100|M.bovis_AF2122/97 -
MMAR_0260|M.marinum_M -
MUL_4854|M.ulcerans_Agy99 -
MAB_0659|M.abscessus_ATCC_1997 -
MLBr_01992|M.leprae_Br4923 -
MAV_0235|M.avium_104 R
MSMEG_0181|M.smegmatis_MC2_155 -