For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
FTADVPAGPIAYDDTGGDGPVLVFGHGLLMDGRQWRKVIPRLPGYRCITPTLPLGAHTVPMRDDADLTQT GVAGILADFLDALDLDDVTLVLNDWGGGQFVISEGRDKRIGRLVLVSCEAFDNFPPKPARPAALLCRFPG GGWVLSRMLGTRLFRHSRRAYGALAKSGIPDAMFDEWFRPAVESAAIRRDLVKFATGSPSRRRLLELSEN LATFDRPVLVVWAREDRMMPLQHADRLVELFSDARKVVVDDSWTLIPEDQPEIMAQLLRDFVG*
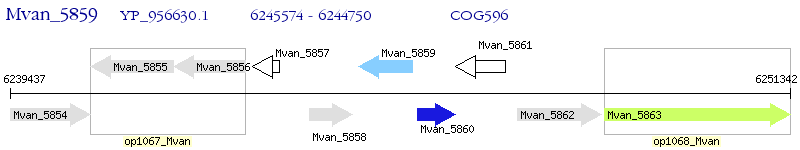
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5859 | - | - | 100% (274) | alpha/beta hydrolase fold |
| M. vanbaalenii PYR-1 | Mvan_4279 | - | 2e-12 | 27.53% (287) | alpha/beta hydrolase fold |
| M. vanbaalenii PYR-1 | Mvan_2912 | - | 9e-10 | 25.61% (289) | alpha/beta hydrolase fold |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0079c | - | 7e-40 | 38.10% (273) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_3236 | - | 1e-135 | 82.05% (273) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv0077c | - | 7e-40 | 38.10% (273) | oxidoreductase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0195c | - | 5e-56 | 44.96% (278) | putative hydrolase, alpha/beta hydrolase fold |
| M. marinum M | MMAR_2774 | - | 3e-38 | 38.61% (259) | oxidoreductase |
| M. avium 104 | MAV_5254 | - | 3e-44 | 40.88% (274) | hydrolase, alpha/beta hydrolase fold family protein |
| M. smegmatis MC2 155 | MSMEG_6763 | - | 1e-133 | 81.85% (270) | oxidoreductase |
| M. thermoresistible (build 8) | TH_3479 | - | 1e-38 | 36.17% (282) | PUTATIVE alpha/beta hydrolase fold |
| M. ulcerans Agy99 | MUL_2973 | - | 9e-38 | 39.43% (246) | oxidoreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_5859|M.vanbaalenii_PYR-1 ---MFTADVPAGPIAYDDTGGDGP---VLVFGHGLLMDGRQWRKVIPRLP
Mflv_3236|M.gilvum_PYR-GCK ---MSTVGVPSGTIAYEDTGGDGP---VMVFGHGLLMDGRQWRKVLPQLP
MSMEG_6763|M.smegmatis_MC2_155 ---MPVIELPSGPIAYEDTGGDGP---VLVFGHGLLMDGRQWRKVIPLLP
MAB_0195c|M.abscessus_ATCC_199 ---MPTFESAAGPIQYRDSG-DGP---PIVFVHGLFVTGTMWRKIVEPLS
TH_3479|M.thermoresistible__bu -MSATTITVDDAAIEYTDVG-GGP---PIVFVHGAYVTGALWDDVVTRLS
Mb0079c|M.bovis_AF2122/97 ---MSTIDISAGTIHYEATGPETG--RPVVFVHGYMMGGQLWRRVSERLA
Rv0077c|M.tuberculosis_H37Rv ---MSTIDISAGTIHYEATGPETG--RPVVFVHGYMMGGQLWRRVSERLA
MAV_5254|M.avium_104 ---MSTIDISAGTIHYEATGPADG--RPVVFVHGYLMGGQLWRQVGDRLA
MMAR_2774|M.marinum_M ---MAQVTLSQATIAYRVLGPENSPHPPVLFVHGILVDERLWSAVAEGLA
MUL_2973|M.ulcerans_Agy99 MAPMAQVTLSQATIAYRVLGPENSPHRPVLFVHGILVDDRLWSAVAEGLA
..* * * ::* ** : * : *.
Mvan_5859|M.vanbaalenii_PYR-1 G--YRCITPTLPLGAHTVPMRDDADLTQTGVAGILADFLDALDLDDVTLV
Mflv_3236|M.gilvum_PYR-GCK E--YRCITPTLPMGAHTSPMRHDADLTETGMANILADFLDALDLDDVTLV
MSMEG_6763|M.smegmatis_MC2_155 G--CRCITPTLPMGAHTAPMRPDADLTEQGVAGILADFLDALDLKDVTLV
MAB_0195c|M.abscessus_ATCC_199 GR-FRCIAPDLPLGAHKTPMNPGVELGPRAVAGLVRDLIVNLDLDDVTVV
TH_3479|M.thermoresistible__bu RG-HRCIAPTWPFGAQRTPVGDSVDLDVVACGRRIIGLLEQLDLRDVTLV
Mb0079c|M.bovis_AF2122/97 GRGLRCIAPTWPLGAHPKPLRPGADQTIGGVAGIVADVLAALELKDVVLV
Rv0077c|M.tuberculosis_H37Rv GRGLRCIAPTWPLGAHPKPLRPGADQTIGGVAGIVADVLAALELKDVVLV
MAV_5254|M.avium_104 GRGLRCIAPTWPMGAHPEPLRRGADRTITGVAAMVAELLSALDLEDVVLV
MMAR_2774|M.marinum_M AMGFRCILPTWPLGSHTIPANEGADLSPAGIAEMIHEFMVALDLADVTLV
MUL_2973|M.ulcerans_Agy99 AMGFRCILPTWPLGSHTIPANEGADLSPAGIAEMIHEFMVALDLADVTLV
*** * *:*:: * ..: . . : .: *:* **.:*
Mvan_5859|M.vanbaalenii_PYR-1 LNDWGGG---QFVISEGRDK-RIGRLVLVSCEAFDNFPPKPARPAALLCR
Mflv_3236|M.gilvum_PYR-GCK LNDWGGG---QFMISEGRDK-RIGRMVLASVTAFDNYPPEPARPAALLCR
MSMEG_6763|M.smegmatis_MC2_155 LNDWGGG---QFLITDGRDA-RIGRLVLASCTAFDNYPPAPARPAAALCR
MAB_0195c|M.abscessus_ATCC_199 ATDTGGAI-TQLLLADGCD--RIGRVVLTPCDSFDNFLPKSIRIFQYWAR
TH_3479|M.thermoresistible__bu ANDSGGGITLSALGILGLDWSRVSRLVFTNCDTFEHFPPKQFAPLVKLCR
Mb0079c|M.bovis_AF2122/97 GNDTGGV---VTQLVAVHYPERLGALVLTSCDAFEHFPPPILKPVILAAK
Rv0077c|M.tuberculosis_H37Rv GNDTGGV---VTQLVAVHYPERLGALVLTSCDAFEHFPPPILKPVILAAK
MAV_5254|M.avium_104 GNDTGGV---VSQLVAVHHRERLGALVLTSCDAFEHFPPPVLRPVILAAK
MMAR_2774|M.marinum_M GNDTGGG---LCQLAIDAHPDLVGRLVLTNCDAFETFPPFPFPAVFTLLR
MUL_2973|M.ulcerans_Agy99 GNDTGGG---LCQLAIDAHPDLVGRLVLTNCDAFETFPPFPFPAVFTLLR
.* ** :. :*:. :*: : * :
Mvan_5859|M.vanbaalenii_PYR-1 FPG--GGWVLSRMLGTRLFRHSRRAYG-ALAKSGIPDAMFDEWFRPAVES
Mflv_3236|M.gilvum_PYR-GCK MPG--GGWVLSRMLGTRFFRHSRRAYG-AMAKSGLPDELFDAWFAPAVEN
MSMEG_6763|M.smegmatis_MC2_155 LPG--GGRLLTKMFGTRFFRHSRLAYG-ALSKAGLPDGLFDEWFRPAVEN
MAB_0195c|M.abscessus_ATCC_199 VPG--TDFVLQPFK----LRLARRALGNTLAKHSIPDAVLGEWVAPLLAQ
TH_3479|M.thermoresistible__bu VAPPLGGAVLRLLATGPGLAFFKRAVT----RNGITGDRDDAIFGGFLSS
Mb0079c|M.bovis_AF2122/97 SAT----LFRAAIQVMRAPAARNR--AYAGLSHHNIDHLTRAWVRPALSN
Rv0077c|M.tuberculosis_H37Rv SAT----LFRAAIQVMRAPAARNR--AYAGLSHHNIDHLTRAWVRPALSN
MAV_5254|M.avium_104 SKA----TFRAVAQAMRAPAVRKR--AFDGLAHSNIDALTELWVRPALSL
MMAR_2774|M.marinum_M GPR----SIKPLFAAMGLRPLRHSPLGYGLLLNKPDAALTASWLQPCRTD
MUL_2973|M.ulcerans_Agy99 RPR----SIKTLFAAMGLRPLRHSPLGFGLLLNKPDAALTASWLQPCRTD
. . .
Mvan_5859|M.vanbaalenii_PYR-1 AAIRRDLVKFATGSPSRRRLLELSENLATFDRPVLVVWAREDRMMPLQHA
Mflv_3236|M.gilvum_PYR-GCK PAIRRDLVKFATGAPPRARLLEMSEQMRTFDRPVLVIWAREDRMMPLEHA
MSMEG_6763|M.smegmatis_MC2_155 AAVRRDLVKFATGAPPRRRLLELSERLRTFTRPVLVVWAREDRMMPLDHA
MAB_0195c|M.abscessus_ATCC_199 QGIRRDVRAFLRGIDNRD-TLAAAEILRGFERPVLLLWPRTLPFFPFAHA
TH_3479|M.thermoresistible__bu AAVRREAVRFSAGLHPQH-TAAAAQAIGKWTKPVLVLWGTADDLFPVSHA
Mb0079c|M.bovis_AF2122/97 PAIAEDLRQLSLSLRTEV-TTAVAARLPEFDKPALIAWSADDVFFALENG
Rv0077c|M.tuberculosis_H37Rv PAIAEDLRQLSLSLRTEV-TTAVAARLPEFDKPALIAWSADDVFFALENG
MAV_5254|M.avium_104 PAVAEDLRRFTLSLRTEV-TTAAAARLYDFDKPTLIAWSADDVFFAQEDG
MMAR_2774|M.marinum_M ARICDDLATLLRHVAATD-LTEVSTRFAQFTKPVTLVWGMGDRCFKPTLA
MUL_2973|M.ulcerans_Agy99 ARICDDLATLLRHVAATD-LTEVSTRFAQFTKPVTLVWGMGDRCFKPTLA
: : : : : : :*. : * : .
Mvan_5859|M.vanbaalenii_PYR-1 DRLVELFSDARKVVVDDSWTLIPEDQPEIMAQLLRDFVG------
Mflv_3236|M.gilvum_PYR-GCK DRLVELYPDATKIVVDDSWTLIPEDQPAAMAQALRDFVSLSPRES
MSMEG_6763|M.smegmatis_MC2_155 DRLVELFPDATKVIVDDSWTLIPEDQPEVMAGALREFVG------
MAB_0195c|M.abscessus_ATCC_199 ERWLRLLPNAKLVEVPDSYTFVSEDQPELMVREIETFVGQRV---
TH_3479|M.thermoresistible__bu QRLAEAFPNSRLRLIEGSSTYVMLDHPSETADAIGTFIAGRS---
Mb0079c|M.bovis_AF2122/97 QRLAATIPRARFEVIEGARTFSMVDSPDRLADQLSTVAVRT----
Rv0077c|M.tuberculosis_H37Rv QRLAATIPRARFEVIEGARTFSMVDSPDRLADQLSTVAVRT----
MAV_5254|M.avium_104 ARLAATIPNARHEVIAGARTFSMVDQPDRLADLLSTVAVRT----
MMAR_2774|M.marinum_M RRMAQLFPDASIIEVPGSKTFVALNEPAAVINAISTVGAHHG---
MUL_2973|M.ulcerans_Agy99 RRMAQLFPDASIIEVPGSKTFVALNEPAAVINAISTVGAHHG---
* . : : .: * : * : .