For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PVIELPSGPIAYEDTGGDGPVLVFGHGLLMDGRQWRKVIPLLPGCRCITPTLPMGAHTAPMRPDADLTEQ GVAGILADFLDALDLKDVTLVLNDWGGGQFLITDGRDARIGRLVLASCTAFDNYPPAPARPAAALCRLPG GGRLLTKMFGTRFFRHSRLAYGALSKAGLPDGLFDEWFRPAVENAAVRRDLVKFATGAPPRRRLLELSER LRTFTRPVLVVWAREDRMMPLDHADRLVELFPDATKVIVDDSWTLIPEDQPEVMAGALREFVG*
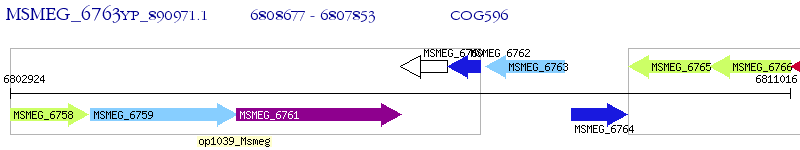
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6763 | - | - | 100% (274) | oxidoreductase |
| M. smegmatis MC2 155 | MSMEG_2517 | - | 7e-12 | 23.75% (299) | hydrolase, alpha/beta fold family protein, putative |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0079c | - | 1e-38 | 37.00% (273) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_3236 | - | 1e-132 | 80.22% (273) | alpha/beta hydrolase fold |
| M. tuberculosis H37Rv | Rv0077c | - | 1e-38 | 37.00% (273) | oxidoreductase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0195c | - | 1e-53 | 43.32% (277) | putative hydrolase, alpha/beta hydrolase fold |
| M. marinum M | MMAR_2774 | - | 8e-40 | 38.04% (276) | oxidoreductase |
| M. avium 104 | MAV_1840 | - | 4e-39 | 36.80% (269) | hydrolase, alpha/beta fold family protein, putative |
| M. thermoresistible (build 8) | TH_3479 | - | 2e-38 | 35.71% (280) | PUTATIVE alpha/beta hydrolase fold |
| M. ulcerans Agy99 | MUL_2973 | - | 2e-39 | 38.27% (277) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_5859 | - | 1e-133 | 81.85% (270) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3236|M.gilvum_PYR-GCK ---MSTVGVPSGTIAYEDTGGDGP---VMVFGHGLLMDGRQWRKVLPQLP
Mvan_5859|M.vanbaalenii_PYR-1 ---MFTADVPAGPIAYDDTGGDGP---VLVFGHGLLMDGRQWRKVIPRLP
MSMEG_6763|M.smegmatis_MC2_155 ---MPVIELPSGPIAYEDTGGDGP---VLVFGHGLLMDGRQWRKVIPLLP
MAB_0195c|M.abscessus_ATCC_199 ---MPTFESAAGPIQYRDSG-DGP---PIVFVHGLFVTGTMWRKIVEPLS
MAV_1840|M.avium_104 ---MASIAVGDVEIDYVDAG-SGP---VVVFVHGAYVTGLLWQDVVDRLS
TH_3479|M.thermoresistible__bu -MSATTITVDDAAIEYTDVG-GGP---PIVFVHGAYVTGALWDDVVTRLS
Mb0079c|M.bovis_AF2122/97 ---MSTIDISAGTIHYEATGPETG--RPVVFVHGYMMGGQLWRRVSERLA
Rv0077c|M.tuberculosis_H37Rv ---MSTIDISAGTIHYEATGPETG--RPVVFVHGYMMGGQLWRRVSERLA
MMAR_2774|M.marinum_M ---MAQVTLSQATIAYRVLGPENSPHPPVLFVHGILVDERLWSAVAEGLA
MUL_2973|M.ulcerans_Agy99 MAPMAQVTLSQATIAYRVLGPENSPHRPVLFVHGILVDDRLWSAVAEGLA
* * * ::* ** : * : *.
Mflv_3236|M.gilvum_PYR-GCK E--YRCITPTLPMGAHTSPMRHDADLTETGMANILADFLDALDLDDVTLV
Mvan_5859|M.vanbaalenii_PYR-1 G--YRCITPTLPLGAHTVPMRDDADLTQTGVAGILADFLDALDLDDVTLV
MSMEG_6763|M.smegmatis_MC2_155 G--CRCITPTLPMGAHTAPMRPDADLTEQGVAGILADFLDALDLKDVTLV
MAB_0195c|M.abscessus_ATCC_199 GR-FRCIAPDLPLGAHKTPMNPGVELGPRAVAGLVRDLIVNLDLDDVTVV
MAV_1840|M.avium_104 DS-HRCIAPTLPFGAQAAPVGAHVDLSVAASGRLIAGLLDALDLSDVTLV
TH_3479|M.thermoresistible__bu RG-HRCIAPTWPFGAQRTPVGDSVDLDVVACGRRIIGLLEQLDLRDVTLV
Mb0079c|M.bovis_AF2122/97 GRGLRCIAPTWPLGAHPKPLRPGADQTIGGVAGIVADVLAALELKDVVLV
Rv0077c|M.tuberculosis_H37Rv GRGLRCIAPTWPLGAHPKPLRPGADQTIGGVAGIVADVLAALELKDVVLV
MMAR_2774|M.marinum_M AMGFRCILPTWPLGSHTIPANEGADLSPAGIAEMIHEFMVALDLADVTLV
MUL_2973|M.ulcerans_Agy99 AMGFRCILPTWPLGSHTIPANEGADLSPAGIAEMIHEFMVALDLADVTLV
*** * *:*:: * .: . . : .: *:* **.:*
Mflv_3236|M.gilvum_PYR-GCK LNDWGGG----QFMISEGRDKRIGRMVLASVTAFDNYPPEPARPAALLCR
Mvan_5859|M.vanbaalenii_PYR-1 LNDWGGG----QFVISEGRDKRIGRLVLVSCEAFDNFPPKPARPAALLCR
MSMEG_6763|M.smegmatis_MC2_155 LNDWGGG----QFLITDGRDARIGRLVLASCTAFDNYPPAPARPAAALCR
MAB_0195c|M.abscessus_ATCC_199 ATDTGGAI--TQLLLADGCD-RIGRVVLTPCDSFDNFLPKSIRIFQYWAR
MAV_1840|M.avium_104 ANDTGGGIVLAALGNASLKWDRVSKLVFTNCDSFEHFPPKSFAPIVRLCA
TH_3479|M.thermoresistible__bu ANDSGGGITLSALGILGLDWSRVSRLVFTNCDTFEHFPPKQFAPLVKLCR
Mb0079c|M.bovis_AF2122/97 GNDTGGV---VTQLVAVHYPERLGALVLTSCDAFEHFPPPILKPVILAAK
Rv0077c|M.tuberculosis_H37Rv GNDTGGV---VTQLVAVHYPERLGALVLTSCDAFEHFPPPILKPVILAAK
MMAR_2774|M.marinum_M GNDTGGG---LCQLAIDAHPDLVGRLVLTNCDAFETFPPFPFPAVFTLLR
MUL_2973|M.ulcerans_Agy99 GNDTGGG---LCQLAIDAHPDLVGRLVLTNCDAFETFPPFPFPAVFTLLR
.* ** :. :*:. :*: : *
Mflv_3236|M.gilvum_PYR-GCK MPGGGWVLSRMLGTRFFRHSRRAYG-AMAKSGLPDELFDAWFAPAVENPA
Mvan_5859|M.vanbaalenii_PYR-1 FPGGGWVLSRMLGTRLFRHSRRAYG-ALAKSGIPDAMFDEWFRPAVESAA
MSMEG_6763|M.smegmatis_MC2_155 LPGGGRLLTKMFGTRFFRHSRLAYG-ALSKAGLPDGLFDEWFRPAVENAA
MAB_0195c|M.abscessus_ATCC_199 VPGTDFVLQPFK----LRLARRALGNTLAKHSIPDAVLGEWVAPLLAQQG
MAV_1840|M.avium_104 LSQS--LGAIALKGLTTAPGLKVFTSAVTLNGVDAARRRAVFGGFLTSPA
TH_3479|M.thermoresistible__bu VAPP--LGGAVLRLLATGPGLAFFKRAVTRNGITGDRDDAIFGGFLSSAA
Mb0079c|M.bovis_AF2122/97 SATLFRAAIQVMRAPAARNR--AYAGLSHHN--IDHLTRAWVRPALSNPA
Rv0077c|M.tuberculosis_H37Rv SATLFRAAIQVMRAPAARNR--AYAGLSHHN--IDHLTRAWVRPALSNPA
MMAR_2774|M.marinum_M GPRSIKPLFAAMGLRPLRHSPLGYGLLLNKP--DAALTASWLQPCRTDAR
MUL_2973|M.ulcerans_Agy99 RPRSIKTLFAAMGLRPLRHSPLGFGLLLNKP--DAALTASWLQPCRTDAR
. . .
Mflv_3236|M.gilvum_PYR-GCK IRRDLVKFATGAPPRARLLEMSEQMRTFDRPVLVIWAREDRMMPLEHADR
Mvan_5859|M.vanbaalenii_PYR-1 IRRDLVKFATGSPSRRRLLELSENLATFDRPVLVVWAREDRMMPLQHADR
MSMEG_6763|M.smegmatis_MC2_155 VRRDLVKFATGAPPRRRLLELSERLRTFTRPVLVVWAREDRMMPLDHADR
MAB_0195c|M.abscessus_ATCC_199 IRRDVRAFLRGIDNRD-TLAAAEILRGFERPVLLLWPRTLPFFPFAHAER
MAV_1840|M.avium_104 VRREAARFTADLKPSH-TLAAVDALKRWDRPVLMAWGTSDKMFPLSHAER
TH_3479|M.thermoresistible__bu VRREAVRFSAGLHPQH-TAAAAQAIGKWTKPVLVLWGTADDLFPVSHAQR
Mb0079c|M.bovis_AF2122/97 IAEDLRQLSLSLRTEV-TTAVAARLPEFDKPALIAWSADDVFFALENGQR
Rv0077c|M.tuberculosis_H37Rv IAEDLRQLSLSLRTEV-TTAVAARLPEFDKPALIAWSADDVFFALENGQR
MMAR_2774|M.marinum_M ICDDLATLLRHVAATD-LTEVSTRFAQFTKPVTLVWGMGDRCFKPTLARR
MUL_2973|M.ulcerans_Agy99 ICDDLATLLRHVAATD-LTEVSTRFAQFTKPVTLVWGMGDRCFKPTLARR
: : : : : :*. : * : . *
Mflv_3236|M.gilvum_PYR-GCK LVELYPDATKIVVDDSWTLIPEDQPAAMAQALRDFVSLSPRES-
Mvan_5859|M.vanbaalenii_PYR-1 LVELFSDARKVVVDDSWTLIPEDQPEIMAQLLRDFVG-------
MSMEG_6763|M.smegmatis_MC2_155 LVELFPDATKVIVDDSWTLIPEDQPEVMAGALREFVG-------
MAB_0195c|M.abscessus_ATCC_199 WLRLLPNAKLVEVPDSYTFVSEDQPELMVREIETFVGQRV----
MAV_1840|M.avium_104 LAETFPNAVVRAIDNSSTYVMLDRPDETAQAIRAFVDPMKDVTR
TH_3479|M.thermoresistible__bu LAEAFPNSRLRLIEGSSTYVMLDHPSETADAIGTFIAGRS----
Mb0079c|M.bovis_AF2122/97 LAATIPRARFEVIEGARTFSMVDSPDRLADQLSTVAVRT-----
Rv0077c|M.tuberculosis_H37Rv LAATIPRARFEVIEGARTFSMVDSPDRLADQLSTVAVRT-----
MMAR_2774|M.marinum_M MAQLFPDASIIEVPGSKTFVALNEPAAVINAISTVGAHHG----
MUL_2973|M.ulcerans_Agy99 MAQLFPDASIIEVPGSKTFVALNEPAAVINAISTVGAHHG----
. : : .: * : * : .