For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDISIIETSGLGDRSYLISHGGSAVVIDPQRDIDRVLALADERATRITHVLETHLHNDYVTGGLELSRTV GAEYVVPAVDGIGYPHRAVTDGDVIDAGPMRFEVLHTPGHTHHHVSYVLRDDTGAVHGVFTGGSMLFGTT GRTDLVAAEDTEELTHAQFHSVRRLADTLPTDTPVYPTHGFGSFCSATPASGDESTIGEQRQSNPALTQD EQSYVDELLAGLSAYPTYYAHMGVINREGPAPVDLTPPAPIDPAELRRRIEAGEWVVDLRNRTAFAAGHL QGTLGFELSGSFVSYFGWLYEWGAPVTLIGESPDQIAEAKRELVRIGVDRLTGSATGDVDTLRAEAPLGS YEVGDFARLARERKTTAVAVLDVRQTNEFDQEHIPGAVSIPLHELPQRLDDVPAGPVWVHCASGYRSSIA ASYLDRAGISVVLIDDDFDNAVKLGLTVSG
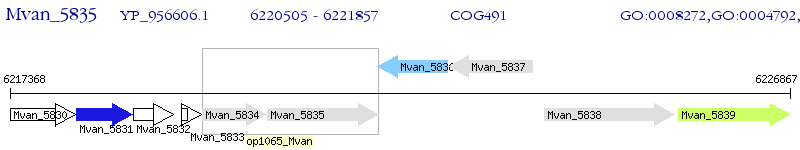
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5835 | - | - | 100% (450) | beta-lactamase domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_4023 | - | 1e-61 | 35.93% (437) | beta-lactamase domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1830 | - | 1e-61 | 35.93% (437) | beta-lactamase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1663c | - | 6e-11 | 25.13% (199) | hypothetical protein Mb1663c |
| M. gilvum PYR-GCK | Mflv_0980 | - | 0.0 | 73.94% (449) | beta-lactamase domain-containing protein |
| M. tuberculosis H37Rv | Rv1637c | - | 6e-11 | 25.13% (199) | hypothetical protein Rv1637c |
| M. leprae Br4923 | MLBr_01391 | - | 1e-11 | 24.04% (208) | hypothetical protein MLBr_01391 |
| M. abscessus ATCC 19977 | MAB_2314c | - | 9e-13 | 28.96% (183) | Beta-lactamase-like |
| M. marinum M | MMAR_2442 | - | 5e-11 | 26.70% (176) | hypothetical protein MMAR_2442 |
| M. avium 104 | MAV_3461 | - | 2e-11 | 29.11% (213) | metallo-beta-lactamase family protein |
| M. smegmatis MC2 155 | MSMEG_5385 | - | 9e-62 | 36.18% (434) | metallo-beta-lactamase family protein |
| M. thermoresistible (build 8) | TH_1234 | - | 2e-62 | 35.47% (437) | metallo-beta-lactamase family protein |
| M. ulcerans Agy99 | MUL_1621 | - | 2e-10 | 26.44% (174) | hypothetical protein MUL_1621 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2442|M.marinum_M ---------------------------------------MTVVDDNYTGH
MUL_1621|M.ulcerans_Agy99 ---------------------------------------MTVVDDNYTGH
Mb1663c|M.bovis_AF2122/97 MLCARTDNHQGTGNVVTSAHMTRANDDDAGAAGIGAVAHMTTVDDNYTGH
Rv1637c|M.tuberculosis_H37Rv MLCARTDNHQGTGNVVTSAHMTRANDDDAGAAGIGAVAHMTTVDDNYTGH
MLBr_01391|M.leprae_Br4923 -----------------------------------MTASIDPVND-YTGH
MAB_2314c|M.abscessus_ATCC_199 -------------------------------------MTSITIDDDYSGH
Mvan_5835|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0980|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5385|M.smegmatis_MC2_155 --------------------------------------------------
TH_1234|M.thermoresistible__bu --------------------------------------------------
MAV_3461|M.avium_104 --------------------------------------------------
MMAR_2442|M.marinum_M VDPGT-VARRTLPGATILKASVGPMDNNAYLVTCSATGQTLLIDA-ANDA
MUL_1621|M.ulcerans_Agy99 VDPGT-VARRTLPGATILKASVGPVDNNAYLVTCSATGQTLLIDA-ANDA
Mb1663c|M.bovis_AF2122/97 VERGK-AARRFLPGATILKASVGPMDNNAYLVTCSATGETLLIDA-ANDA
Rv1637c|M.tuberculosis_H37Rv VERGK-AARRFLPGATILKASVGPMDNNAYLVTCSATGETLLIDA-ANDA
MLBr_01391|M.leprae_Br4923 VDPGT-AARRALPGATILKVSVGPMDNNAYLVTCSATGETLLIDA-ANDA
MAB_2314c|M.abscessus_ATCC_199 VEPGSGVARRSVDGATIIKASVGPMDNNAYIVTCTKTGKSLLIDA-ANDA
Mvan_5835|M.vanbaalenii_PYR-1 --------------MDISIIETSGLGDRSYLISHG--GSAVVIDP-QRDI
Mflv_0980|M.gilvum_PYR-GCK ----------MSSAIDVSIIETSGLGDRSYLISAD--GTAVVVDP-QRDI
MSMEG_5385|M.smegmatis_MC2_155 --------------MKFTQYYLDCLSHASYLIADETTGRAVVVDP-QRDV
TH_1234|M.thermoresistible__bu --------------MKFIQYYLDCLSHASYLIADDKTGRAVVVDP-QRDV
MAV_3461|M.avium_104 --------------MLITGFPAGMLACNCYVLAQRPGTDAVIVDPGQRAM
. . : .*::: ::::*. .
MMAR_2442|M.marinum_M EVLIDLIRRYAPKIALIVTSHQHFDHWQALQAVAEATGAPTAAHEIDAEP
MUL_1621|M.ulcerans_Agy99 EVLIDLIRRYAPKIALIVTSHQHFDHWQALQAVAEATGAPTAAHEIDAEP
Mb1663c|M.bovis_AF2122/97 EVLIDLVRRYAPKLALIVTSHQHFDHWQALQAVAAATGAPTAAHPIDADP
Rv1637c|M.tuberculosis_H37Rv EVLIDLVRRYAPKLALIVTSHQHFDHWQALQAVAAATGAPTAAHPIDADP
MLBr_01391|M.leprae_Br4923 EVLIDLVQRYAPKLSLIVTSHQHFDHWQALEAVVAATGAPTAVHELDAEP
MAB_2314c|M.abscessus_ATCC_199 ERLATLVDESAPNIELIVTTHQHFDHWQALEAIVGKTQSPSAAHDLDAGP
Mvan_5835|M.vanbaalenii_PYR-1 DRVLALADERATRITHVLETHLHNDYVTGGLELSRTVGAEYVVPAVDGIG
Mflv_0980|M.gilvum_PYR-GCK DRVLDLARERGVRISHVLETHIHNDYVTGGLELSRVTGAEYVVPAGDDVG
MSMEG_5385|M.smegmatis_MC2_155 AGYLADAEEFGYTIELVIETHFHADFLSGHLELAKATGAKIVYSSVAETE
TH_1234|M.thermoresistible__bu AEYLSDAEKYGYTIELVIETHFHADFLSGHLELAEATGAKIVYSSVAETE
MAV_3461|M.avium_104 GPLRRILDRHRLTPAAVLLTHGHIDHIWSAQKVSDTYGCPTYIHPEDRFM
. :: :* * *. . : .
MMAR_2442|M.marinum_M LPVKPD----------------------RLLANGDRVQIGELSFDVIHLR
MUL_1621|M.ulcerans_Agy99 LPVKPD----------------------RLLANGDRVQIGELSFDVIHLR
Mb1663c|M.bovis_AF2122/97 LPVKPD----------------------RLLTHGDSVRIGELTFDVIHLR
Rv1637c|M.tuberculosis_H37Rv LPVKPD----------------------RLLTHGDSVRIGELTFDVIHLR
MLBr_01391|M.leprae_Br4923 LPVRPD----------------------RLLTNGDCVQIGELTFDVIHLR
MAB_2314c|M.abscessus_ATCC_199 LPVRPN----------------------TLLAGGDSLDIGDLHFDVIHLR
Mvan_5835|M.vanbaalenii_PYR-1 YPHRA-------------------------VTDGDVIDAGPMRFEVLHTP
Mflv_0980|M.gilvum_PYR-GCK YERRA-------------------------VSDGDTVDAGPVRLQVMHTP
MSMEG_5385|M.smegmatis_MC2_155 FESMG-------------------------VADGERYSLGEVTLEFRHTP
TH_1234|M.thermoresistible__bu FESMG-------------------------VADGERYSLGDVTLEFLHTP
MAV_3461|M.avium_104 LKDPIYGLGPRLAQLAAGAFFREPRQVVELDRDGDKLDLGSVTVNVDHTP
*: * : .:. *
MMAR_2442|M.marinum_M GHTPGSIALALD----GAATGGVTQLFTGDCLFPGGVGKT-------WQP
MUL_1621|M.ulcerans_Agy99 GHTPGSIALALD----GAATGGVTQLFTGDCLFPGGVGKT-------WQP
Mb1663c|M.bovis_AF2122/97 GHTPGSIALALG----GPVTGGVTQLFTGDCLFPGGVGKT-------WQP
Rv1637c|M.tuberculosis_H37Rv GHTPGSIALALG----GPVTGGVTQLFTGDCLFPGGVGKT-------WQP
MLBr_01391|M.leprae_Br4923 GHTPGSIALALDGATSGPKTGRVTQLFTGDCLFPGGVGKT-------WQR
MAB_2314c|M.abscessus_ATCC_199 GHTPGSVALAFT-------AAGVTHLFTGDSLFPGGPGRT-------TNP
Mvan_5835|M.vanbaalenii_PYR-1 GHTHHHVSYVLR--DDTG---AVHGVFTGGSMLFGTTGRTDLVA--AEDT
Mflv_0980|M.gilvum_PYR-GCK GHTHHHVSYVLR--DAGGSGDSVLGVFTGGSMLHGTTGRTDLLG--DEHT
MSMEG_5385|M.smegmatis_MC2_155 GHTPESMSVVVY--EHSND-DVPYGVLTGDALFIGDVGRPDLLASIGFTR
TH_1234|M.thermoresistible__bu GHTPESLSIVVY--EHADD-EIPYGVLTGDTLFIGDVGRPDLLASIGYTR
MAV_3461|M.avium_104 GHTRGSMCFRVS--------SDKQVVFTGDTLFERSVGRT-------DLF
*** :. . ::**. :: *:.
MMAR_2442|M.marinum_M GDFTQLVEDVSSRVFDVYPDSTVVYPGHGDDTVLGV-----ERPHLDEWR
MUL_1621|M.ulcerans_Agy99 GDFTQLVEDVSSRVFNVYPDSTVVYPGHGDDTALGF-----ERPHLDEWR
Mb1663c|M.bovis_AF2122/97 ADFTQLLDDVTTRVFDVYADSTVIYPGHGDDTELGA-----ERPSLSEWR
Rv1637c|M.tuberculosis_H37Rv ADFTQLLDDVTTRVFDVYADSTVIYPGHGDDTELGA-----ERPSLSEWR
MLBr_01391|M.leprae_Br4923 SDFTQLLDDVTTRVFDVYADTTAIYPGHGDDTVLGA-----ERPSLAEWR
MAB_2314c|M.abscessus_ATCC_199 EEFTSLMDDLEGLVFGRYPDDTVVYPGHGDDTTLGS-----ERPHLAEWR
Mvan_5835|M.vanbaalenii_PYR-1 EELTHAQFHSVRRLADTLPTDTPVYPTHGFGSFCSATPASGDESTIGEQR
Mflv_0980|M.gilvum_PYR-GCK QELSHAQFHSVRRLAAELPDTAEVYPTHGFGSFCSATPASGDSSTIADQR
MSMEG_5385|M.smegmatis_MC2_155 EELAEKLYDSLRNKLMTLPDATRVYPAHGAGSACGKNLSTERWSTIGEQK
TH_1234|M.thermoresistible__bu EELADKLYDSLHHKLMTLPDATRVYPAHGAGSACGKNLSTELWSTIGEQR
MAV_3461|M.avium_104 GGSGRDLLTSIVDKLLVLDDDTVVLPGHGNATTIGA------ERRLNPFL
: : * ** : . :
MMAR_2442|M.marinum_M ERGW----------------------------------------------
MUL_1621|M.ulcerans_Agy99 ERGW----------------------------------------------
Mb1663c|M.bovis_AF2122/97 ARGW----------------------------------------------
Rv1637c|M.tuberculosis_H37Rv ARGW----------------------------------------------
MLBr_01391|M.leprae_Br4923 ERGW----------------------------------------------
MAB_2314c|M.abscessus_ATCC_199 KRGW----------------------------------------------
Mvan_5835|M.vanbaalenii_PYR-1 QSNPALTQ-DEQSYVDELLAGLSAYPTYYAHMGVINREGPAPVDLT-PPA
Mflv_0980|M.gilvum_PYR-GCK QTNPALTQ-DEQSYVDELIAGLGAYPAYYAHMGVINTEGPDPVDLS-VPE
MSMEG_5385|M.smegmatis_MC2_155 QTNYALRAPDLATFMALVTAGQPAAPSYFVYDAILNRKDRDLLDETEIPA
TH_1234|M.thermoresistible__bu ETNYALRAPDKATFIELVTEGQPPAPGYFVYDAILNRKNRGLLDETALPP
MAV_3461|M.avium_104 EGL-----------------------------------------------
MMAR_2442|M.marinum_M --------------------------------------------------
MUL_1621|M.ulcerans_Agy99 --------------------------------------------------
Mb1663c|M.bovis_AF2122/97 --------------------------------------------------
Rv1637c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01391|M.leprae_Br4923 --------------------------------------------------
MAB_2314c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_5835|M.vanbaalenii_PYR-1 PIDPAELRRRIEAGEWVVDLRNRTAFAAGHLQGTLGFELSGSFVSYFGWL
Mflv_0980|M.gilvum_PYR-GCK PVDPDELRRRIEGGEWVVDLRNRTAFAAGHLGGTLGFELSDNFVTYLGWL
MSMEG_5385|M.smegmatis_MC2_155 AMRYEQVNQALRDGAILVDGRSPEEFATGHLRQAINIGLQGRYAEFAGSV
TH_1234|M.thermoresistible__bu VLSYDEMLAAVERGAVLVDGRSPEEFAQGHLRGAINIGLDGRYAEFAGSV
MAV_3461|M.avium_104 --------------------------------------------------
MMAR_2442|M.marinum_M --------------------------------------------------
MUL_1621|M.ulcerans_Agy99 --------------------------------------------------
Mb1663c|M.bovis_AF2122/97 --------------------------------------------------
Rv1637c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01391|M.leprae_Br4923 --------------------------------------------------
MAB_2314c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_5835|M.vanbaalenii_PYR-1 YEWGAPVTLIGESPDQIAEAKRELVRIGVDRLTGSATG--DVDTLRAEAP
Mflv_0980|M.gilvum_PYR-GCK YSWGSPLTLIGDDVDQIADARRELARIGVDNLTGSATG--DIHDLAEGTP
MSMEG_5385|M.smegmatis_MC2_155 VPADVDIVLFTE-PGEELEAKNRLARIGFDRVIGHLDRPFEVMLAHPDDV
TH_1234|M.thermoresistible__bu LPADVDIVLFTE-PGDELEAKNRLARIGFDRVIGHLDRPFEVMMAHPDRV
MAV_3461|M.avium_104 --------------------------------------------------
MMAR_2442|M.marinum_M --------------------------------------------------
MUL_1621|M.ulcerans_Agy99 --------------------------------------------------
Mb1663c|M.bovis_AF2122/97 --------------------------------------------------
Rv1637c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01391|M.leprae_Br4923 --------------------------------------------------
MAB_2314c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_5835|M.vanbaalenii_PYR-1 LGSYEVGDFARLARERKTTAVAVLDVRQTNEFDQEHIPGAVSIPLHELPQ
Mflv_0980|M.gilvum_PYR-GCK LRSYRVADFAALADAMQAQAPAVLDVRQSGEFADGHIAGAVNIPLHELSR
MSMEG_5385|M.smegmatis_MC2_155 QMASRLTATAFDQRTAEVADLQIVDVRNPGEVADGAIPHAITIPVGQLPA
TH_1234|M.thermoresistible__bu QVASRLTVKAFDQRAAELPDLQIVDVRNPGEVALGAIPGAIAIPVGQLPD
MAV_3461|M.avium_104 --------------------------------------------------
MMAR_2442|M.marinum_M --------------------------------------------------
MUL_1621|M.ulcerans_Agy99 --------------------------------------------------
Mb1663c|M.bovis_AF2122/97 --------------------------------------------------
Rv1637c|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_01391|M.leprae_Br4923 --------------------------------------------------
MAB_2314c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_5835|M.vanbaalenii_PYR-1 RLDDVP-AGPVWVHCASGYRSSIAASYLDRAGIS-VVLIDDDFDNAVKLG
Mflv_0980|M.gilvum_PYR-GCK RLDEVP-DGEVWVHCASGYRSSIAASMIDRGDRT-VVLVDDAFDQAEKTG
MSMEG_5385|M.smegmatis_MC2_155 RLGELDPAKPTVVYCAGGYRSSVAASVLRRNGFADVSDILGGYGAWRDAR
TH_1234|M.thermoresistible__bu RLTELDPTRPTVVYCAGGYRSSVAASLLRRSGFTDVSDILGGYGAWVDAR
MAV_3461|M.avium_104 --------------------------------------------------
MMAR_2442|M.marinum_M -----
MUL_1621|M.ulcerans_Agy99 -----
Mb1663c|M.bovis_AF2122/97 -----
Rv1637c|M.tuberculosis_H37Rv -----
MLBr_01391|M.leprae_Br4923 -----
MAB_2314c|M.abscessus_ATCC_199 -----
Mvan_5835|M.vanbaalenii_PYR-1 LTVSG
Mflv_0980|M.gilvum_PYR-GCK LVS--
MSMEG_5385|M.smegmatis_MC2_155 QGV--
TH_1234|M.thermoresistible__bu QVA--
MAV_3461|M.avium_104 -----