For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SEPPGGLRGRTALVTGGAGGIGAACARALAEQGVTVTVADIDDVGAKSVAQEIGGKAWALDLLDVEALEG LQLETDILVNNAGVQSIHEIAEFPPEKFRSMMRLMVEAPFLLIRAAVPHMYRNEFGRVINISSIHGLRAS PYKAAYVTAKHALEGLSKVTALEGGPHGVTSNCINPGYVRTPLVTRQIADQARTHNIGEDQVLSDILLKE SAVKRLVEPEEVGALAAWLASPVAGMVTGASYTMDGGWSAR*
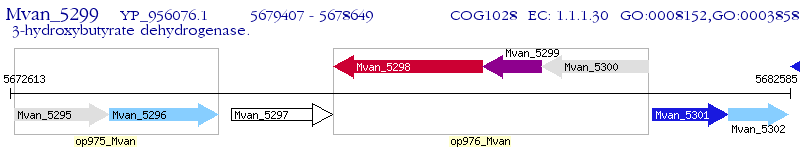
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_5299 | - | - | 100% (252) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_3000 | - | 1e-31 | 33.98% (256) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_4209 | - | 2e-28 | 33.86% (254) | short-chain dehydrogenase/reductase SDR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2025 | fabG3 | 2e-31 | 36.33% (256) | 20-beta-hydroxysteroid dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1472 | - | 1e-125 | 90.61% (245) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv2002 | fabG3 | 2e-31 | 36.33% (256) | 20-beta-hydroxysteroid dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 2e-20 | 29.51% (244) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_4680c | - | 1e-31 | 32.68% (254) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_2981 | fabG3_1 | 1e-33 | 37.94% (253) | 20-beta-hydroxysteroid dehydrogenase FabG3_1 |
| M. avium 104 | MAV_5136 | - | 7e-33 | 34.92% (252) | 3-alpha-(or 20-beta)-hydroxysteroid dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6026 | - | 1e-104 | 75.51% (245) | 3-hydroxybutyrate dehydrogenase |
| M. thermoresistible (build 8) | TH_4644 | - | 1e-84 | 77.08% (192) | PUTATIVE secreted protein |
| M. ulcerans Agy99 | MUL_1037 | fabG3 | 3e-33 | 34.92% (252) | 20-beta-hydroxysteroid dehydrogenase FabG3 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2025|M.bovis_AF2122/97 --------------------------------------------------
Rv2002|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2981|M.marinum_M --------------------------------------------------
MAV_5136|M.avium_104 --------------------------------------------------
MUL_1037|M.ulcerans_Agy99 --------------------------------------------------
MAB_4680c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01807|M.leprae_Br4923 --------------------------------------------------
Mvan_5299|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1472|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_6026|M.smegmatis_MC2_155 --------------------------------------------------
TH_4644|M.thermoresistible__bu LNRPAFGLAVITPLVFAGAVGGSGPLQFPHESDTAVTPLAAVSPEPVDRS
Mb2025|M.bovis_AF2122/97 --------------------------------------------------
Rv2002|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2981|M.marinum_M --------------------------------------------------
MAV_5136|M.avium_104 --------------------------------------------------
MUL_1037|M.ulcerans_Agy99 --------------------------------------------------
MAB_4680c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01807|M.leprae_Br4923 --------------------------------------------------
Mvan_5299|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1472|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_6026|M.smegmatis_MC2_155 --------------------------------------------------
TH_4644|M.thermoresistible__bu GPTVVALQKKPAPFHFVESTVAAPPPAAVVNTPGTLGIPAMALRAYRNAE
Mb2025|M.bovis_AF2122/97 -------------------------------------------MSGR---
Rv2002|M.tuberculosis_H37Rv -------------------------------------------MSGR---
MMAR_2981|M.marinum_M -------------------------------------------MAGR---
MAV_5136|M.avium_104 --------------------------------------------MGR---
MUL_1037|M.ulcerans_Agy99 --------------------------------------------MGR---
MAB_4680c|M.abscessus_ATCC_199 -------------------------------------------MNGNS-A
MLBr_01807|M.leprae_Br4923 ---------------------------------MTDAAAKVSAAPGRGKP
Mvan_5299|M.vanbaalenii_PYR-1 -------------------------------------------MSEP-PG
Mflv_1472|M.gilvum_PYR-GCK -------------------------------------------MSESGMS
MSMEG_6026|M.smegmatis_MC2_155 ------------------------------------------------MT
TH_4644|M.thermoresistible__bu RMMAHAQPECGVSWNLLAGIGRIESMHAYGGATDANGTAIRPILGPLLDG
Mb2025|M.bovis_AF2122/97 -LIGKVALVSG-------------------------GARGMG--------
Rv2002|M.tuberculosis_H37Rv -LIGKVALVSG-------------------------GARGMG--------
MMAR_2981|M.marinum_M -LTGKVALVSG-------------------------GARGMG--------
MAV_5136|M.avium_104 -VDGKVALISG-------------------------GARGMG--------
MUL_1037|M.ulcerans_Agy99 -VDGKVALISG-------------------------GARGMG--------
MAB_4680c|M.abscessus_ATCC_199 LLQGKNVIVTG-------------------------GARGLG--------
MLBr_01807|M.leprae_Br4923 AFVSRSILVTG-------------------------GNRGIG--------
Mvan_5299|M.vanbaalenii_PYR-1 GLRGRTALVTG-------------------------GAGGIG--------
Mflv_1472|M.gilvum_PYR-GCK DLRGRTAVVTG-------------------------GAGGIG--------
MSMEG_6026|M.smegmatis_MC2_155 DLAGRKALVTG-------------------------AASGIG--------
TH_4644|M.thermoresistible__bu SLPGNEIIVQSRTADRVTYARALGPMQFLPGTWARFASDGDGDGKADVQN
. .. :: . . * *
Mb2025|M.bovis_AF2122/97 -----ASHVRAMVAEGAKVVFGD---------------------------
Rv2002|M.tuberculosis_H37Rv -----ASHVRAMVAEGAKVVFGD---------------------------
MMAR_2981|M.marinum_M -----ASHVRALVAEGAHVVLGD---------------------------
MAV_5136|M.avium_104 -----AEHARLLAAEGAKVVIGD---------------------------
MUL_1037|M.ulcerans_Agy99 -----ASHARLLVQEGAKVVIGD---------------------------
MAB_4680c|M.abscessus_ATCC_199 -----AAFSRHIVSQGGRVVIAD---------------------------
MLBr_01807|M.leprae_Br4923 -----LAVARRLVADGHRVAVT----------------------------
Mvan_5299|M.vanbaalenii_PYR-1 -----AACARALAEQGVTVT------------------------------
Mflv_1472|M.gilvum_PYR-GCK -----AACAKALAERGVTVT------------------------------
MSMEG_6026|M.smegmatis_MC2_155 -----AACVRELAARGAAVT------------------------------
TH_4644|M.thermoresistible__bu LFDSTLAAARYLCSGGMNLRDPSHVMQALLRYNNSVAYAHNVMGWAAAYA
: : * :
Mb2025|M.bovis_AF2122/97 ---------------------ILDEEGKAVAAELADAAR-------YVHL
Rv2002|M.tuberculosis_H37Rv ---------------------ILDEEGKAVAAELADAAR-------YVHL
MMAR_2981|M.marinum_M ---------------------ILDDEGRAVAAELGDAAR-------YVHL
MAV_5136|M.avium_104 ---------------------ILDDEGKAVADEIGDSVR-------YVHL
MUL_1037|M.ulcerans_Agy99 ---------------------ILDEEGKALAEEIGDAAR-------YVHL
MAB_4680c|M.abscessus_ATCC_199 ---------------------VLDGDGRQLAAELGDAAR-------FTHL
MLBr_01807|M.leprae_Br4923 ----------------------------HRASVVPDWFF-------GVEC
Mvan_5299|M.vanbaalenii_PYR-1 ---------------------VADIDDVGAKSVAQEIGG-----------
Mflv_1472|M.gilvum_PYR-GCK ---------------------VADIDEVGAKTVAEEIGG-----------
MSMEG_6026|M.smegmatis_MC2_155 ---------------------IADIDAERADALAAEVGG-----------
TH_4644|M.thermoresistible__bu TGVHPVDLPPLTGPVPSLNSHVSGYDGLGPGSPLNAMGLSPGDPLAVLPL
Mb2025|M.bovis_AF2122/97 DVTQPAQWTAAVDTAVTAFGGLHVLVNNAGILNIGTIEDYALTEWQRILD
Rv2002|M.tuberculosis_H37Rv DVTQPAQWTAAVDTAVTAFGGLHVLVNNAGILNIGTIEDYALTEWQRILD
MMAR_2981|M.marinum_M DVTQPEQWTAAVDTAVNEFGGLHVLVNNAGILNIGTIEDYALSEWQRILD
MAV_5136|M.avium_104 DVTQPDQWDAAVETAVGEFGKLNVLVNNAGTVALGPLKSFDLAKWQKVID
MUL_1037|M.ulcerans_Agy99 DVTQPDQWEAAVATAVDEFGKLDVLVNNVGIVALGQLKKFDLGKWQKVID
MAB_4680c|M.abscessus_ATCC_199 DVTDAEQWRELIDTTLAGFGTITGLVNNAGISSAAMVADEPLEHFRKVIE
MLBr_01807|M.leprae_Br4923 DVTDNDAIGAAFKAVEEHQGPVEVLVANAGITSDMLIMRMSEEQFQKVID
Mvan_5299|M.vanbaalenii_PYR-1 --KAWALDLLDVEALEGLQLETDILVNNAGVQSIHEIAEFPPEKFRSMMR
Mflv_1472|M.gilvum_PYR-GCK --KAWALDLLDVAALESLQLETDILVNNAGVQNISEIAEFPPEKFRSMMA
MSMEG_6026|M.smegmatis_MC2_155 --TAWAVDLLDVAALEDLELDVDILVNNAGVQSINPIEEFPPERFRMLMT
TH_4644|M.thermoresistible__bu IDGXXXLDLLDRSALHDLRLDIDILVNNAGVQHVDPVTEFPPDQFRDMLT
: ** *.* : .:: ::
Mb2025|M.bovis_AF2122/97 VNLTGVFLGIRAVVKPMKEAGRGSIINISSIEGLAGTVACHGYTATKFAV
Rv2002|M.tuberculosis_H37Rv VNLTGVFLGIRAVVKPMKEAGRGSIINISSIEGLAGTVACHGYTATKFAV
MMAR_2981|M.marinum_M INVTGVFLGIRAAVKPMKEAGRGSIINISSIEGLAGTIASHGYTVSKFAV
MAV_5136|M.avium_104 VNLTGTFLGMRVAVEPMIAAGGGSIINISSIEGLRGAPMVHPYVASKWGV
MUL_1037|M.ulcerans_Agy99 VNLTGTFLGMRAAVEPMTAAGSGSIINVSSIEGLRGAPAVHPYVASKWAV
MAB_4680c|M.abscessus_ATCC_199 INLVGVFIGMQAVIPAMRAHGCGSIVNISSAAGLVGLALTSGYGASKWGV
MLBr_01807|M.leprae_Br4923 VNLTGVFRVAKLASRNMIKQRFGRYIFMGSVVGLSGVPGQANYAASKSGL
Mvan_5299|M.vanbaalenii_PYR-1 LMVEAPFLLIRAAVPHMYRNEFGRVINISSIHGLRASPYKAAYVTAKHAL
Mflv_1472|M.gilvum_PYR-GCK LMVEAPFLLIRAALPHMYRNEFGRVINISSIHGLRASPFKVAYVTAKHAL
MSMEG_6026|M.smegmatis_MC2_155 LMVESPFLLIRAALPHMYRQRFGRIVNISSVHGLRASEYKVGYVTAKHAL
TH_4644|M.thermoresistible__bu LMVEAPFLLIRAALPYMFERGFGRIVNISSVHGLRASEYKAAYVTAKHAL
: : . * : . * * : :.* ** . * .:* .:
Mb2025|M.bovis_AF2122/97 RGLTKSTALELGPSGIRVNSIHPGLVKTPMTDWV----------------
Rv2002|M.tuberculosis_H37Rv RGLTKSTALELGPSGIRVNSIHPGLVKTPMTDWV----------------
MMAR_2981|M.marinum_M RGLTKSTALELGPSGIRVNSIHPGLVKTPMTEWV----------------
MAV_5136|M.avium_104 RGLAKSAALELAPHNIRVNSVHPGFIRTPMTKHL----------------
MUL_1037|M.ulcerans_Agy99 RGLTKSAALELAPLNIRVNSIHPGFIRTPMTANL----------------
MAB_4680c|M.abscessus_ATCC_199 RGLSKVAAIELGRERIRVNSVHPGVIYTPMTAKEGFREG-----------
MLBr_01807|M.leprae_Br4923 IGMARSLARELGSRSITANVVAPGFIDTEMTRAMDSKYQ-----------
Mvan_5299|M.vanbaalenii_PYR-1 EGLSKVTALEGGPHGVTSNCINPGYVRTPLVTRQIADQARTHNIGEDQVL
Mflv_1472|M.gilvum_PYR-GCK EGLSKVTALEGGPHGVTSNCVNPGYVRTPLVTKQIADQARTHGIPEDQVV
MSMEG_6026|M.smegmatis_MC2_155 EGLSKVTALEGGPHQVTSNCINPGYVRTPLVTKQIADQARIHGIPEDDVV
TH_4644|M.thermoresistible__bu EGLSKVTALEGGPHGVTSNCVNPGYVRTPLVTRQIADQARAHGISEGEVL
*::: * * . : * : ** : * :.
Mb2025|M.bovis_AF2122/97 -PEDIFQTALGRAAEPVEVSNLVVYLASDESSYSTGAEFVVDGGTVAGLA
Rv2002|M.tuberculosis_H37Rv -PEDIFQTALGRAAEPVEVSNLVVYLASDESSYSTGAEFVVDGGTVAGLA
MMAR_2981|M.marinum_M -PEDLFQTALGRAAEPMEVSNLVVYLASDESSYSTGAEFVVDGGTVAGLA
MAV_5136|M.avium_104 -PDDMVTVPLGRPAESREVSTFVLFLASDESSYATGSEFVMDGGLVTDVP
MUL_1037|M.ulcerans_Agy99 -PDDMVTIPLGRPAESREVSTFVVFLASDDASYATGSEFVMDGGLVTDVP
MAB_4680c|M.abscessus_ATCC_199 -EGNMPIAAMGRVGNADEVAGAVAYLLSDAASYVTGAELAVDGGWTAGPT
MLBr_01807|M.leprae_Br4923 -EAALQAIPLGRIGAVEEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH-
Mvan_5299|M.vanbaalenii_PYR-1 SDILLKESAVKRLVEPEEVGALAAWLASPVAGMVTGASYTMDGGWSAR--
Mflv_1472|M.gilvum_PYR-GCK SDILLKESAVKRLVGPEEVGSLVAWLASPAAGMVTGASYTMDGGWSAR--
MSMEG_6026|M.smegmatis_MC2_155 AQVLLKESAIKQLVEPEEVAALVGWLASPSARMVTGASYTMDGGWSAR--
TH_4644|M.thermoresistible__bu EQILLKESAIKRLIEPDEVASLVGWLASPDAGMVTGASYTMDGGWSAR--
: .: : *:. . :* * : :*: :***
Mb2025|M.bovis_AF2122/97 HNDFGAVEVSSQPEWVT
Rv2002|M.tuberculosis_H37Rv HNDFGAVEVSSQPEWVT
MMAR_2981|M.marinum_M HKDFSAVDVAAQPEWVT
MAV_5136|M.avium_104 HKQF-------------
MUL_1037|M.ulcerans_Agy99 HKQF-------------
MAB_4680c|M.abscessus_ATCC_199 LTTITGQ----------
MLBr_01807|M.leprae_Br4923 -----------------
Mvan_5299|M.vanbaalenii_PYR-1 -----------------
Mflv_1472|M.gilvum_PYR-GCK -----------------
MSMEG_6026|M.smegmatis_MC2_155 -----------------
TH_4644|M.thermoresistible__bu -----------------