For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VDDRYIDTTVLVTGAASGIGRAIAFAAAREGARLILGDITVDRLADAAEELRARGAQVVHARCDITSTAD LESLVQRGSHELGPVDVAFANAGVLGSPGDVWSYSEDEFTRILDINVTGTWRTVRAVLPAMIERRRGIIV ATASAAGLIGPAGLPAYVASKHAVVGLVKSTAMNVAAHGIRVNALCPHMVDTPMLDKVSHEIPGLRESLD QQTPLGRVATSEEVARSALWLGSDESSFVTGHALLVDGGLVAQ
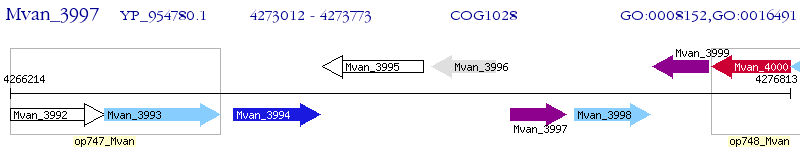
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3997 | - | - | 100% (253) | short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_1975 | fabG | 1e-38 | 36.36% (242) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. vanbaalenii PYR-1 | Mvan_1286 | fabG | 5e-38 | 36.94% (268) | 3-ketoacyl-(acyl-carrier-protein) reductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2025 | fabG3 | 8e-38 | 40.16% (249) | 20-beta-hydroxysteroid dehydrogenase |
| M. gilvum PYR-GCK | Mflv_4369 | fabG | 2e-41 | 38.02% (242) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. tuberculosis H37Rv | Rv2002 | fabG3 | 8e-38 | 40.16% (249) | 20-beta-hydroxysteroid dehydrogenase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 1e-28 | 29.27% (246) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_3844c | - | 8e-36 | 36.68% (259) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_4033 | fabG | 2e-37 | 35.15% (239) | 3-oxoacyl- |
| M. avium 104 | MAV_1572 | - | 9e-38 | 34.71% (242) | short chain dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_2206 | fabG | 1e-38 | 38.02% (242) | 3-ketoacyl-(acyl-carrier-protein) reductase |
| M. thermoresistible (build 8) | TH_3434 | - | 9e-39 | 36.80% (269) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_3898 | fabG | 1e-37 | 35.15% (239) | 3-ketoacyl-(acyl-carrier-protein) reductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4369|M.gilvum_PYR-GCK -------MSLLTG--------QTAVITGGAQGLGFAIAQRFVDEGARVVL
MSMEG_2206|M.smegmatis_MC2_155 ---------MLTG--------QTAVVTGGAQGLGLAIAKRFISEGARVVL
MMAR_4033|M.marinum_M -----MSVSLLNG--------QIAVVTGGAQGLGLAIAERFVSEGARVVL
MUL_3898|M.ulcerans_Agy99 -----MCGAHRDRG------YVYPVVTGGAQGLGLAIAERFVSEGARVVL
MAV_1572|M.avium_104 -------MSLLSG--------QTAVITGGAQGLGFAIAERFVAEGARVVL
MLBr_01807|M.leprae_Br4923 MTDAAAKVSAAPGRGKPAFVSRSILVTGGNRGIGLAVARRLVADGHRVAV
MAB_3844c|M.abscessus_ATCC_199 -----MADQPLAG--------KVAFVTGAARGQGRQHAVRLAQAGADVVA
TH_3434|M.thermoresistible__bu ----------------------VAFITGAARGQGRAHAIRLAEEGADIIA
Mvan_3997|M.vanbaalenii_PYR-1 ------MDDRYID--------TTVLVTGAASGIGRAIAFAAAREGARLIL
Mb2025|M.bovis_AF2122/97 ------MSGRLIG--------KVALVSGGARGMGASHVRAMVAEGAKVVF
Rv2002|M.tuberculosis_H37Rv ------MSGRLIG--------KVALVSGGARGMGASHVRAMVAEGAKVVF
.::*. * * . * :
Mflv_4369|M.gilvum_PYR-GCK GDVN--------------LEATQEAAEKLGGADVAHAVRCDVTSSAEVDA
MSMEG_2206|M.smegmatis_MC2_155 GDLN--------------SEATEAAVEELGGSEVAAAVRCDVTSSADVDA
MMAR_4033|M.marinum_M GDVN--------------LEATEAAAKQLGGGEVAVAVRCDVTKADEVET
MUL_3898|M.ulcerans_Agy99 GDVN--------------LEATEAAAKQLGGGEVAVAVRCDVTKADEVET
MAV_1572|M.avium_104 GDVN--------------LEATQTAAKQLGGDQVALAVRCDVTKSSEVET
MLBr_01807|M.leprae_Br4923 THR-----------------ASVVPDWFFG-------VECDVTDNDAIGA
MAB_3844c|M.abscessus_ATCC_199 IDACAPVSEYAGYEAATPEDLKETVRLVEAAGRKIIAEQADVRNGAALQS
TH_3434|M.thermoresistible__bu IDVCGPVSDTITYPPATEDDLAETVRAVEATGRKVLARTADIRDLAAQQQ
Mvan_3997|M.vanbaalenii_PYR-1 GD-------------ITVDRLADAAEELRARGAQVVHARCDITSTADLES
Mb2025|M.bovis_AF2122/97 GD--------------ILDEEGKAVAAELADAARYVH--LDVTQPAQWTA
Rv2002|M.tuberculosis_H37Rv GD--------------ILDEEGKAVAAELADAARYVH--LDVTQPAQWTA
. . *: .
Mflv_4369|M.gilvum_PYR-GCK LVAAAVDTFGALDIMVNNAGIT-RDATMRKMTEDDFDKVIAVHLKGTWNG
MSMEG_2206|M.smegmatis_MC2_155 LVQAAVERFGGLDIMVNNAGIT-RDATLRKMTEEQFDQVIAVHLKGTWNG
MMAR_4033|M.marinum_M LLQTALERFGGLDIMVNNAGIT-RDATMRKMTEEQFDQVIDVHLKGTWNG
MUL_3898|M.ulcerans_Agy99 LLQAALERFGGLDIMVNNAGIT-RDATMRKMTEEQFDQVIDVHLKGTWNG
MAV_1572|M.avium_104 LIQTAVERFGGLDIMVNNAGIT-RDATMRKMTEEQFDQVIAVHLKGTWNG
MLBr_01807|M.leprae_Br4923 AFKAVEEHQGPVEVLVANAGIT-SDMLIMRMSEEQFQKVIDVNLTGVFRV
MAB_3844c|M.abscessus_ATCC_199 VVEEAVAQFGHIDILIANAGVL-SWGRLWEMSDQQWEDIIDTNLTGTWKT
TH_3434|M.thermoresistible__bu IVAEGMERFGRLDIVVANAGVL-SWGRLWEMSEEQWDTVIDVNLNGTWRT
Mvan_3997|M.vanbaalenii_PYR-1 LVQRGSHELGPVDVAFANAGVLGSPGDVWSYSEDEFTRILDINVTGTWRT
Mb2025|M.bovis_AF2122/97 AVDTAVTAFGGLHVLVNNAGIL-NIGTIEDYALTEWQRILDVNLTGVFLG
Rv2002|M.tuberculosis_H37Rv AVDTAVTAFGGLHVLVNNAGIL-NIGTIEDYALTEWQRILDVNLTGVFLG
. * :.: . ***: : : :: :: ::.*.:
Mflv_4369|M.gilvum_PYR-GCK LRAAAAIMRENKRG-VIVNMSSISGKVGMIGQTNYSAAKAGIVGMTKAAS
MSMEG_2206|M.smegmatis_MC2_155 TKAAAAIMRENKRG-AIVNMSSISGKVGLIGQTNYSAAKAGIVGMTKAAA
MMAR_4033|M.marinum_M IRLAAAIMRENKRG-AIVNMSSVSGKVGMVGQTNYSAAKAGIVGMTKAAA
MUL_3898|M.ulcerans_Agy99 IRLAAAIMRENKRG-AIVNMSSVSGKVGMVGQTNYSAAKAGIVGMTKAAA
MAV_1572|M.avium_104 TRLAAAIMRENKRG-AIINMSSVSGKVGMVGQTNYSAAKAGIVGMTKAAA
MLBr_01807|M.leprae_Br4923 AKLASRNMIKQRFG-RYIFMGSVVGLSGVPGQANYAASKSGLIGMARSLA
MAB_3844c|M.abscessus_ATCC_199 VKAVVPTMIEAGNGGSIVIVSSVSGLKGTPGNGAYVASKFGLTGLTKSLT
TH_3434|M.thermoresistible__bu IRAVVPAMIEAGNGGSIIIVSSSAGLKATPGNGHYAASKHGLVALTNALA
Mvan_3997|M.vanbaalenii_PYR-1 VRAVLPAMIERRRG-IIVATASAAGLIGPAGLPAYVASKHAVVGLVKSTA
Mb2025|M.bovis_AF2122/97 IRAVVKPMKEAGRG-SIINISSIEGLAGTVACHGYTATKFAVRGLTKSTA
Rv2002|M.tuberculosis_H37Rv IRAVVKPMKEAGRG-SIINISSIEGLAGTVACHGYTATKFAVRGLTKSTA
: . * : * : .* * . . * *:* .: .:..: :
Mflv_4369|M.gilvum_PYR-GCK KELAYLGVRVNAIQPGLIRSAMTE-------AMPQRIWDSKVAEVPLG--
MSMEG_2206|M.smegmatis_MC2_155 KELAYLGVRVNAIQPGLIRSAMTE-------AMPQRIWDEKLAEIPMG--
MMAR_4033|M.marinum_M KELAHVGVRVNAIAPGLIRSAMTE-------AMPQRIWDQKLAEVPMG--
MUL_3898|M.ulcerans_Agy99 KELAHVGVRVNAIAPGLIRSAMTE-------AMPQRIWDQKLAEVPMG--
MAV_1572|M.avium_104 KELAYLGVRVNAIAPGLIRSAMTE-------AMPQRIWDSKVAEVPMG--
MLBr_01807|M.leprae_Br4923 RELGSRSITANVVAPGFIDTEMTR-------AMDSKYQEAALQAIPLG--
MAB_3844c|M.abscessus_ATCC_199 IELAEYGIRVNSIHPYGVTTPMITGDAMMKILAEHPNYLPSIAPMPLSPT
TH_3434|M.thermoresistible__bu IEVGEYGIRVNSIHPYSIDTPMIERDAMMAVFAKHPNYLHSFAPMPLHPV
Mvan_3997|M.vanbaalenii_PYR-1 MNVAAHGIRVNALCPHMVDTPMLD-----KVSHEIPGLRESLDQQTPLG-
Mb2025|M.bovis_AF2122/97 LELGPSGIRVNSIHPGLVKTPMTD-------WVPEDIFQTALG-------
Rv2002|M.tuberculosis_H37Rv LELGPSGIRVNSIHPGLVKTPMTD-------WVPEDIFQTALG-------
::. .: .* : * : : * .
Mflv_4369|M.gilvum_PYR-GCK ---------RAGEPDEVAKVALFLASDLSSYMTGTVLEVTGGRHL-----
MSMEG_2206|M.smegmatis_MC2_155 ---------RAGEPDEVAKVALFLASDLSSYMTGTVLEVTGGRHV-----
MMAR_4033|M.marinum_M ---------RAGEPSEVASVALFLACDLSSYMTGTVLDVTGGRFI-----
MUL_3898|M.ulcerans_Agy99 ---------RAGEPSEVASVALFLACDLSSYMTGTVLDVTGGRFI-----
MAV_1572|M.avium_104 ---------RAGEPSEVASVALFLASDMSSYMTGTVMEITGGRHL-----
MLBr_01807|M.leprae_Br4923 ---------RIGAVEEIAGAVSFLASEDAGYISGAVIPVDGGLGMGH---
MAB_3844c|M.abscessus_ATCC_199 ---------KMLEPDEISDVVLWLASDASGTLAGAQVPVDKGHLTY----
TH_3434|M.thermoresistible__bu NREGDGGRQEFMTPEEVADVVAWLAGDGSATLSGSQIAVDRGVMKY----
Mvan_3997|M.vanbaalenii_PYR-1 ---------RVATSEEVARSALWLGSDESSFVTGHALLVDGGLVAQ----
Mb2025|M.bovis_AF2122/97 ---------RAAEPVEVSNLVVYLASDESSYSTGAEFVVDGGTVAGLAHN
Rv2002|M.tuberculosis_H37Rv ---------RAAEPVEVSNLVVYLASDESSYSTGAEFVVDGGTVAGLAHN
. *:: . :*. : :. :* . : *
Mflv_4369|M.gilvum_PYR-GCK ---------------
MSMEG_2206|M.smegmatis_MC2_155 ---------------
MMAR_4033|M.marinum_M ---------------
MUL_3898|M.ulcerans_Agy99 ---------------
MAV_1572|M.avium_104 ---------------
MLBr_01807|M.leprae_Br4923 ---------------
MAB_3844c|M.abscessus_ATCC_199 ---------------
TH_3434|M.thermoresistible__bu ---------------
Mvan_3997|M.vanbaalenii_PYR-1 ---------------
Mb2025|M.bovis_AF2122/97 DFGAVEVSSQPEWVT
Rv2002|M.tuberculosis_H37Rv DFGAVEVSSQPEWVT