For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ERHALEARSSGVASGDHDDTDARTPRTMSHPHPDRSADDAQPRWGGAALLRPGVLAFAGSIGATDAHAHH AVQIITAATPLTVTDEQGASHTGTKVVVPTDAAHRIRAGAQDGTVVFLEPESAPGRAAHLRAIDSGWAIS SALAFTAHRPLATVVAELVAHLSPATFEHDVPARHSSVDHALRLLPALLAAGPVRGTELAARVGLSASRF THLFTAQVGIPLRRYVLWSRLRIAITLVQGGDDLTGAAHGAGFADSAHLTRTTREMFGLPPSVLSRHVSW DLDPSS*
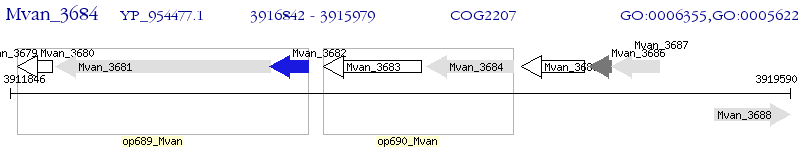
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3684 | - | - | 100% (287) | helix-turn-helix domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_3649 | - | e-101 | 73.97% (242) | helix-turn-helix domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1839 | - | 1e-16 | 33.19% (229) | helix-turn-helix domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3863 | - | 4e-05 | 27.56% (127) | AraC family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_2796 | - | 1e-30 | 34.73% (239) | helix-turn-helix domain-containing protein |
| M. tuberculosis H37Rv | Rv3833 | - | 4e-05 | 27.56% (127) | AraC family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0066c | - | 1e-32 | 38.33% (240) | AraC family transcriptional regulator |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_0221 | - | 4e-06 | 27.05% (244) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_2039 | - | 6e-15 | 44.34% (106) | putative transcriptional regulator |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3863|M.bovis_AF2122/97 -----------------------------------MSENSHHRLATTSLT
Rv3833|M.tuberculosis_H37Rv -----------------------------------MSENSHHRLATTSLT
Mflv_2796|M.gilvum_PYR-GCK ----------------------------------MRTRHTRDVTWLGEAL
MAB_0066c|M.abscessus_ATCC_199 ------------------------------------------MVWTGAAV
Mvan_3684|M.vanbaalenii_PYR-1 MERHALEARSSGVASGDHDDTDARTPRTMSHPHPDRSADDAQPRWGGAAL
MSMEG_2039|M.smegmatis_MC2_155 --------------------------------MPTPIPTPLPRTCYPAVW
MAV_0221|M.avium_104 -------------------------MYRPSPPLAEHIEYFGYWRGDEALG
Mb3863|M.bovis_AF2122/97 LPPG--------ARIERHRHPSHQIVYPSAGAVSVTTHAGTWITPVNRAI
Rv3833|M.tuberculosis_H37Rv LPPG--------ARIERHRHPSHQIVYPSAGAVSVTTHAGTWITPVNRAI
Mflv_2796|M.gilvum_PYR-GCK IEPGKLVFRGQLGSAHAHGHAALQIVTIDEGAAFLVDADGRGLWAP--AA
MAB_0066c|M.abscessus_ATCC_199 IEPGRLVYRGALGDTHRHTHAAVQIAIALDGTLQLTDGADRRVSTR--AA
Mvan_3684|M.vanbaalenii_PYR-1 LRPGVLAFAGSIGATDAHAHHAVQIITAAT-PLTVTDEQGASHTGT--KV
MSMEG_2039|M.smegmatis_MC2_155 LWPGQGLYSGPALGLQPHSGSVWCLVVGVDAPLTVDVG-GHQISAR--TA
MAV_0221|M.avium_104 VHTSRALPRGAVTAIIDVGGRTDLGFYASDARTPLTVPPAFAAGAGATAY
: .. :
Mb3863|M.bovis_AF2122/97 WIPAGCWHQHKFHGHTQFHGVALDPQRYRGGPATPTVLAVNPLMRELIIA
Rv3833|M.tuberculosis_H37Rv WIPAGCWHQHKFHGHTQFHGVALDPQRYRGGPATPTVLAVNPLMRELVIA
Mflv_2796|M.gilvum_PYR-GCK IIPAGAEHSIEAEACQGMM-LYLEPTSVIGSAVTALFRNTNRNDVREWVR
MAB_0066c|M.abscessus_ATCC_199 VIPAGAQHTVHGGTSTGLM-IYLEPTCFEGRALGALFDNAARDDATAWCA
Mvan_3684|M.vanbaalenii_PYR-1 VVPTDAAHRIRAGAQDGTV-VFLEPESAPGRAAHLRAIDSGWAISSALAF
MSMEG_2039|M.smegmatis_MC2_155 LIPPRVTHHLAMIG--PLVSCYLDPASHRTASCRRQFDDIVHGIGIGHHR
MAV_0221|M.avium_104 VVRVAPAHTVMTIHFRPAGALAFLGSPLSDLEDALVGLEDLWGRDAALLR
: * :
Mb3863|M.bovis_AF2122/97 CSQADRTDTDEHHRMLAVLQDQLPTTSIREPLWVPSPTDRRLRHACALIA
Rv3833|M.tuberculosis_H37Rv CSQADRTDTDEHHRMLAVLQDQLPTTSIREPLWVPSPTDRRLRHACALIA
Mflv_2796|M.gilvum_PYR-GCK LGKKLVNINVVEQQHLSSAASEVIAYLVGSEGQPRCAVHPGVEAAVDLLP
MAB_0066c|M.abscessus_ATCC_199 AASELSDI-----QDDSSSAESVIARLVG--DVHASEPHESVREAIRLLP
Mvan_3684|M.vanbaalenii_PYR-1 TAHRPLAT---------VVAELVAHLSPATFEHDVPARHSSVDHALRLLP
MSMEG_2039|M.smegmatis_MC2_155 EDLLLNPP---------ADDSSAVRWLDTAAPSVIHDIDPRIELAAKQIR
MAV_0221|M.avium_104 EQLIDAGS---------PPRRVALLQAFLVRRMRRNAVWPPARLAPVLRG
*
Mb3863|M.bovis_AF2122/97 DNLTQ-PLTLQQIGGRIGVSQRTLSRLFSDELGMTFPQWRTQLRLQHALV
Rv3833|M.tuberculosis_H37Rv DNLTQ-PLTLQQIGGRIGVSQRTLSRLFSDELGMTFPQWRTQLRLQHALV
Mflv_2796|M.gilvum_PYR-GCK GIIEG-PVKLSDVARTVHLSADRLGRLFAHEVGMSIPAYVRWCRLIRAME
MAB_0066c|M.abscessus_ATCC_199 GMLTG-PVRLAEVARVVHLSADRLGRMFARDTGMSFPAYVRWARLIRTIE
Mvan_3684|M.vanbaalenii_PYR-1 ALLAAGPVRGTELAARVGLSASRFTHLFTAQVGIPLRRYVLWSRLRIAIT
MSMEG_2039|M.smegmatis_MC2_155 DDPAT-VVAAAELAATAGLSESRFLHLFRAELGTSLRRYRMWCRLLRAGA
MAV_0221|M.avium_104 ADLDP-SMRVSKAQELSGLSRKRFAALFRCEVGLSPKAYLRVLRLQAALR
: . :* : :* : * . : ** :
Mb3863|M.bovis_AF2122/97 LLAERHDVTSVASECGWATPSAFIDTYRQAFGHTPGQAAKPMAATRLTRL
Rv3833|M.tuberculosis_H37Rv LLAERHDVTSVASECGWATPSAFIDTYRQAFGHTPGQAAKPMAATRLTRL
Mflv_2796|M.gilvum_PYR-GCK VVRDGGTITDAAHAAGFSDSAHANRVFHEMFGLAPIDARRGVRLT-----
MAB_0066c|M.abscessus_ATCC_199 VARTGGTLTDAAHAAGFSDSSHANRAFHEMFGITPVELHRSVQLS-----
Mvan_3684|M.vanbaalenii_PYR-1 LVQGGDDLTGAAHGAGFADSAHLTRTTREMFGLPPSVLSRHVSWDLDPSS
MSMEG_2039|M.smegmatis_MC2_155 LLATGHDLTTAAVDSGFASPSHLADRFKSTFGLSASQLLATGLQIRVP--
MAV_0221|M.avium_104 ALDTPARGATIAADLGYFDQAHFVREFRAFTGATPTQYARRRSSMPGHVE
: * *: : : * ..
Mb3863|M.bovis_AF2122/97 RRARDRR
Rv3833|M.tuberculosis_H37Rv RRARDRR
Mflv_2796|M.gilvum_PYR-GCK -------
MAB_0066c|M.abscessus_ATCC_199 -------
Mvan_3684|M.vanbaalenii_PYR-1 -------
MSMEG_2039|M.smegmatis_MC2_155 -------
MAV_0221|M.avium_104 LAR----