For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTRAGGDTTGAPRWGGTALLRPGVLALTGSIGTTDVHAHHAVQILTATTPLTVRDGHGIGHRGTKVVVPA DAPHRIEVGAQEGTVVFLEPESLPGRSAHLRAVHSGWTVAPVLTSTRRRTLAAVVDELVEHLAPVLTGDG ATPRHPAVDDALRLLPSLVAAGPVSGTDLAAQVGLSVSRLTHLFTDQVGIPLRRYVLWSRLRLAVMRVQA GDDLTGAAHGAGFADSAHLTRTTREMFGLAPSVLSRHVSWDLEAGL
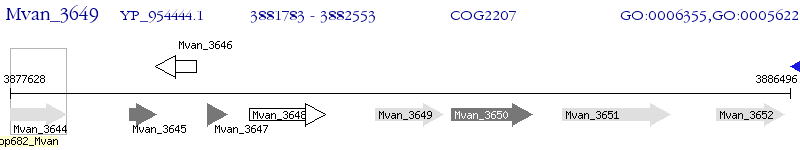
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_3649 | - | - | 100% (256) | helix-turn-helix domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_3684 | - | e-101 | 73.97% (242) | helix-turn-helix domain-containing protein |
| M. vanbaalenii PYR-1 | Mvan_1839 | - | 1e-15 | 31.17% (231) | helix-turn-helix domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0552 | - | 9e-05 | 28.97% (214) | hypothetical protein Mb0552 |
| M. gilvum PYR-GCK | Mflv_2796 | - | 1e-32 | 36.36% (242) | helix-turn-helix domain-containing protein |
| M. tuberculosis H37Rv | Rv0538 | - | 9e-05 | 28.97% (214) | hypothetical protein Rv0538 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0066c | - | 3e-29 | 34.73% (239) | AraC family transcriptional regulator |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_4981 | - | 3e-06 | 33.33% (102) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_2039 | - | 1e-12 | 40.00% (100) | putative transcriptional regulator |
| M. thermoresistible (build 8) | TH_2340 | - | 7e-08 | 25.38% (260) | transcriptional regulatory protein |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2796|M.gilvum_PYR-GCK --------------------------------------------------
MAB_0066c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_3649|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb0552|M.bovis_AF2122/97 MDVALGVAVTDRVARLALVDSAAPGTVIDQFVLDVAEHPVEVLTETVVGT
Rv0538|M.tuberculosis_H37Rv MDVALGVAVTDRVARLALVDSAAPGTVIDQFVLDVAEHPVEVLTETVVGT
MSMEG_2039|M.smegmatis_MC2_155 --------------------------------------------------
TH_2340|M.thermoresistible__bu --------------------------------------------------
MAV_4981|M.avium_104 --------------------------------------------------
Mflv_2796|M.gilvum_PYR-GCK --------------------------------------------------
MAB_0066c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_3649|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb0552|M.bovis_AF2122/97 DRSLAGENHRLVATRLCWPDQAKADELQHALQDSGVHDVAVISEAQAATA
Rv0538|M.tuberculosis_H37Rv DRSLAGENHRLVATRLCWPDQAKADELQHALQDSGVHDVAVISEAQAATA
MSMEG_2039|M.smegmatis_MC2_155 --------------------------------------------------
TH_2340|M.thermoresistible__bu --------------------------------------------------
MAV_4981|M.avium_104 ---------------------------------MADSSATPRVVVIVVFD
Mflv_2796|M.gilvum_PYR-GCK -------------------------------------------MRTRHTR
MAB_0066c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_3649|M.vanbaalenii_PYR-1 ----------------------------------------MTRAGGDTTG
Mb0552|M.bovis_AF2122/97 LVGAAHAGSAVLLVGDETATLSVVGDPDAPPTMVAVAPVAGADATSTVDT
Rv0538|M.tuberculosis_H37Rv LVGAAHAGSAVLLVGDETATLSVVGDPDAPPTMVAVAPVAGADATSTVDT
MSMEG_2039|M.smegmatis_MC2_155 -----------------------------------------MPTPIPTPL
TH_2340|M.thermoresistible__bu --------------------------------------VTVSALSARNLA
MAV_4981|M.avium_104 DVTMLDVAGAGEVFAEANRFGADYLLKIASVDGRDVTTSIGTRLGVTDAV
Mflv_2796|M.gilvum_PYR-GCK DVTWLGEALIEPGKLVFRG------QLGSAHAHGHAALQIVTIDEGAAFL
MAB_0066c|M.abscessus_ATCC_199 -MVWTGAAVIEPGRLVYRG------ALGDTHRHTHAAVQIAIALDGTLQL
Mvan_3649|M.vanbaalenii_PYR-1 APRWGGTALLRPGVLALTG------SIGTTDVHAHHAVQILTATT-PLTV
Mb0552|M.bovis_AF2122/97 LMARLGDQALAPGDVFLVGRSAEHTTVLADQLRAASTMRVQTPDDPTFAL
Rv0538|M.tuberculosis_H37Rv LMARLGDQALAPGDVFLVGRSAEHTTVLADQLRAASTMRVQTPDDPTFAL
MSMEG_2039|M.smegmatis_MC2_155 PRTCYPAVWLWPGQGLYSG------PALGLQPHSGSVWCLVVGVDAPLTV
TH_2340|M.thermoresistible__bu SSAKPPQIDLRRGGRALGGSYLYEGDKLQTGWHSHDVHQIEYAIGGVVEV
MAV_4981|M.avium_104 SSIESADTVLVAGSDNLPRRPIDPELVEAVKSVADRTRRLASICTGSFIL
: * . : :
Mflv_2796|M.gilvum_PYR-GCK VDADGRGLW--APAAIIPAGAEHS--------------------------
MAB_0066c|M.abscessus_ATCC_199 TDGADRRVS--TRAAVIPAGAQHT--------------------------
Mvan_3649|M.vanbaalenii_PYR-1 RDGHGIGHR--GTKVVVPADAPHR--------------------------
Mb0552|M.bovis_AF2122/97 ARGAAMAAG--AATMAHPALVADATTSLPPAEAGQSGSEGEQLAYSQASD
Rv0538|M.tuberculosis_H37Rv ARGAAMAAG--AATMAHPALVADATTSLPRAEAGQSGSEGEQLAYSQASD
MSMEG_2039|M.smegmatis_MC2_155 DVG-GHQIS--ARTALIPPRVTHH--------------------------
TH_2340|M.thermoresistible__bu QTRTGHYLLPPQQAAWLPVGLEHS--------------------------
MAV_4981|M.avium_104 GQAGLLDGRR-ATTHWHDVRLFAR--------------------------
Mflv_2796|M.gilvum_PYR-GCK ------------IEAEACQGMMLYLEPTSVIGSAVTALFRNTNRNDVREW
MAB_0066c|M.abscessus_ATCC_199 ------------VHGGTSTGLMIYLEPTCFEGRALGALFDNAARDDATAW
Mvan_3649|M.vanbaalenii_PYR-1 ------------IEVGAQEGTVVFLEPESLPGRSAHLRAVHSGWTVAPVL
Mb0552|M.bovis_AF2122/97 YELLPVDEYEEHDEYGAAADRSAPLSRRSLLIGNAVVAFAVIGFASLAVA
Rv0538|M.tuberculosis_H37Rv YELLPVDEYEEHDEYGAAADRSAPLSRRSLLIGNAVVAFAVIGFASLAVA
MSMEG_2039|M.smegmatis_MC2_155 -------------LAMIGPLVSCYLDPASHRTASCRRQFDDIVHGIGIGH
TH_2340|M.thermoresistible__bu -----------ATMNPDVKTVAVMFDPQLVPDAGNRARILAVSPLIREMM
MAV_4981|M.avium_104 ----------AFPEITVEPDALFVCDGDVFTSAGISSGIDLALALVEMDY
.
Mflv_2796|M.gilvum_PYR-GCK VRLGKKLVNINVVEQQHLSSAASEVIAYLVGSEGQPRCAVHPG-------
MAB_0066c|M.abscessus_ATCC_199 CAAASELSDI-----QDDSSSAESVIARLVG--DVHASEPHES-------
Mvan_3649|M.vanbaalenii_PYR-1 TSTRRRTLAA------VVDELVEHLAPVLTG---DGATPRHPA-------
Mb0552|M.bovis_AF2122/97 VAVTIRPTAASKPVEGHQNAQPGKFMPLLPTQQQAPVPPPPPDDPTAGFQ
Rv0538|M.tuberculosis_H37Rv VAVTIRPTAASKPVEGHQNAQPGKFMPLLPTQQQAPVPPPPPDDPTAGFQ
MSMEG_2039|M.smegmatis_MC2_155 HREDLLLNPP---------ADDSSAVRWLDTAAPSVIHDIDPR-------
TH_2340|M.thermoresistible__bu IYALRWPIDR-----PHGDEISDTFFRTLAHLVSEALDHEAPLSLP----
MAV_4981|M.avium_104 GTELVRAVAR---------WLVVYLKRAGGQSQFSALVEAEPPPRS----
Mflv_2796|M.gilvum_PYR-GCK -----VEAAVDLLPGIIEG-PVKLSDVARTVHLSADRLGRLFAHEVGMSI
MAB_0066c|M.abscessus_ATCC_199 -----VREAIRLLPGMLTG-PVRLAEVARVVHLSADRLGRMFARDTGMSF
Mvan_3649|M.vanbaalenii_PYR-1 -----VDDALRLLPSLVAAGPVSGTDLAAQVGLSVSRLTHLFTDQVGIPL
Mb0552|M.bovis_AF2122/97 GGTIPAVQNVVPRPGTSPGVGGTPASPAPEAPAVPGVVPAPVPIPVPIII
Rv0538|M.tuberculosis_H37Rv GGTIPAVQNVVPRPGTSPGVGGTPASPAPEAPAVPGVVPAPVPIPVPIII
MSMEG_2039|M.smegmatis_MC2_155 -----IELAAKQIR-DDPATVVAAAELAATAGLSESRFLHLFRAELGTSL
TH_2340|M.thermoresistible__bu -TSDHPIVAAAMAYTKEHLQSVTAEEVSRAVAVSERTLRRQFRQATGISW
MAV_4981|M.avium_104 -----PLRKVTAAISADPAADHSVKSLSARASLSTRQLTRLFQSELGTTP
. : . . .
Mflv_2796|M.gilvum_PYR-GCK PAYVRWCRLIRAMEVVRD-GGTITDAAHAAGFSDSAHANRVFHEMFGLAP
MAB_0066c|M.abscessus_ATCC_199 PAYVRWARLIRTIEVART-GGTLTDAAHAAGFSDSSHANRAFHEMFGITP
Mvan_3649|M.vanbaalenii_PYR-1 RRYVLWSRLRLAVMRVQA-GDDLTGAAHGAGFADSAHLTRTTREMFGLAP
Mb0552|M.bovis_AF2122/97 PPFPGWQPGMPTIPTAPP-TTPVTTSATTPPTTPPTTPVTTPPTTPPTTP
Rv0538|M.tuberculosis_H37Rv PPFPGWQPGMPTIPTAPP-TTPVTTSATTPPTTPPTTPVTTPPTTPPTTP
MSMEG_2039|M.smegmatis_MC2_155 RRYRMWCRLLRAGALLAT-GHDLTTAAVDSGFASPSHLADRFKSTFGLSA
TH_2340|M.thermoresistible__bu RTYLLHARMLKAMALLAAPDQSVQETASAVGFDSLSAFTRAFTQFCGETP
MAV_4981|M.avium_104 ARYVEMVRIDAARAALDA-GRSVAESARLAGFGSAETLRRVFVEHLGLSP
: : : :* :.
Mflv_2796|M.gilvum_PYR-GCK IDARRGVRLT----------------------------------------
MAB_0066c|M.abscessus_ATCC_199 VELHRSVQLS----------------------------------------
Mvan_3649|M.vanbaalenii_PYR-1 SVLSRHVSWDLEAGL-----------------------------------
Mb0552|M.bovis_AF2122/97 VTTPPTTPPTTPVTTPPTTVAPTTVAPTTVAPTTVAPTTVAPATATPTTV
Rv0538|M.tuberculosis_H37Rv VTTPPTTPPTTPVTTPPTTVAPTTVAPTTVAPTTVAPTTVAPATATPTTV
MSMEG_2039|M.smegmatis_MC2_155 SQLLATGLQIRVP-------------------------------------
TH_2340|M.thermoresistible__bu SAYRHRISTDPHSRPTSADPIG----------------------------
MAV_4981|M.avium_104 KAYRDRFRTAARG-------------------------------------
Mflv_2796|M.gilvum_PYR-GCK --------------------------------------------------
MAB_0066c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_3649|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb0552|M.bovis_AF2122/97 APQPTQQPTQQPTQQMPTQQQTVAPQTVAPAPQPPSGGRNGSGGGDLFGG
Rv0538|M.tuberculosis_H37Rv APQPTQQPTQQPTQQMPTQQQTVAPQTVAPAPQPPSGGRNGSGGGDLFGG
MSMEG_2039|M.smegmatis_MC2_155 --------------------------------------------------
TH_2340|M.thermoresistible__bu --------------------------------------------------
MAV_4981|M.avium_104 --------------------------------------------------
Mflv_2796|M.gilvum_PYR-GCK -
MAB_0066c|M.abscessus_ATCC_199 -
Mvan_3649|M.vanbaalenii_PYR-1 -
Mb0552|M.bovis_AF2122/97 F
Rv0538|M.tuberculosis_H37Rv F
MSMEG_2039|M.smegmatis_MC2_155 -
TH_2340|M.thermoresistible__bu -
MAV_4981|M.avium_104 -