For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSPDQRGVRALTVQGWQYLVLSVMGFVVLTGGIMVAVLLNRTDQVSGQLIERIQPARVAALQMQAAIRDQ ETAVRGYLITADLRFLKPYDEGRAAEQQASADIRQLEQDRPDLLDDLDAIEQASGSWRTNYAEPVVADVR PGDPGSVDPELTAIGQADFDALRVLFDTQNENLAQARDADIAELERMRTLRNAVLLAMAVTFAVTVVALA VLLRRALVRPLSRLAASCRRIAEGNFGERIVPQGPRDIRLISLDVEHMRQRIVEELEASRAATEALDEQA EELRRSNAELEQFAYVASHDLQEPLRKVASFCQLLEKRYGDQLDERGVEYIGFAVDGAKRMQILINDLLT FSRVGRLNATETEVDLDEALDNALSNLATAVEESDAVIERPVGGLPTVKGDPTLLTMLWQNLIGNAVKFR REGVAPRIVIDVAQSGDDDAQNWSFSLADNGIGIAAEFTDKVFVIFQRLHGRDAYSGTGIGLALCKKIVE HHGGTIWIDSSYTEGTRFRFTLPMTLDDTATPLLEGSTP
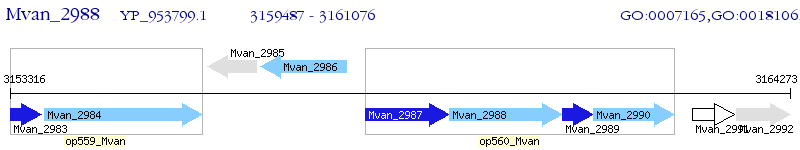
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2988 | - | - | 100% (529) | multi-sensor signal transduction histidine kinase |
| M. vanbaalenii PYR-1 | Mvan_5174 | - | 4e-23 | 27.91% (326) | integral membrane sensor signal transduction histidine kinase |
| M. vanbaalenii PYR-1 | Mvan_1421 | - | 4e-18 | 25.36% (280) | ATPase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0781 | phoR | 1e-24 | 28.97% (359) | two component system response sensor kinase membrane |
| M. gilvum PYR-GCK | Mflv_3269 | - | 0.0 | 80.27% (527) | multi-sensor signal transduction histidine kinase |
| M. tuberculosis H37Rv | Rv0758 | phoR | 1e-24 | 28.97% (359) | two component system response sensor kinase membrane |
| M. leprae Br4923 | MLBr_02124 | - | 4e-17 | 26.13% (333) | sensor histidine kinase |
| M. abscessus ATCC 19977 | MAB_0674 | - | 7e-23 | 29.83% (352) | putative sensor histidine kinase PhoR |
| M. marinum M | MMAR_1264 | - | 0.0 | 63.53% (521) | two-component system membrane associated sensor kinase |
| M. avium 104 | MAV_4241 | - | 1e-174 | 61.38% (523) | two-component sensor protein |
| M. smegmatis MC2 155 | MSMEG_6130 | - | 0.0 | 68.25% (504) | two-component sensor protein |
| M. thermoresistible (build 8) | TH_2175 | - | 0.0 | 62.92% (542) | two-component sensor protein |
| M. ulcerans Agy99 | MUL_2624 | - | 0.0 | 64.11% (521) | two component system membrane associated sensor kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2988|M.vanbaalenii_PYR-1 ------MSPDQRGVRALTVQGWQYLVLSVMGFVVLTGGIMVAVLLNRTDQ
Mflv_3269|M.gilvum_PYR-GCK ------MSPDQRGVRALTVQGWQYVVLAVMGLVVLVGGIAVSVLLHRTDQ
MSMEG_6130|M.smegmatis_MC2_155 --------------MRLTVQGWQNLVLSVMGVVVFTGAIAGAVLVGRTDT
TH_2175|M.thermoresistible__bu ---VTPAATGERRGSRLTVQGWQNLVLSGMGVIVLAGAITGWLLLHRTDD
MMAR_1264|M.marinum_M MTPEPPIRPRTRRRIALTVRGWQALVLSIMGVMVLAGTVAGAVLLNRTDD
MUL_2624|M.ulcerans_Agy99 MTPEPPIRPRTRRRIALTVRGWQALVLSIMGVMVLAGTVAGAVLLNRTDG
MAV_4241|M.avium_104 MT-----APRPLRLTQLTVRGWLALVLWIMGTTVLIGALVGAVLLHRTDQ
Mb0781|M.bovis_AF2122/97 -----------------------MARHLRGRLPLRVRLVAATLILVATGL
Rv0758|M.tuberculosis_H37Rv -----------------------MARHLRGRLPLRVRLVAATLILVATGL
MAB_0674|M.abscessus_ATCC_1997 MAWDTYCASRDERLREEQLALLVAVPGVRRGVPLRVSLVVGTLVLVALGL
MLBr_02124|M.leprae_Br4923 --------------------MNILSRIFARTPSLRTRVVVATAIGAAIPV
: : :
Mvan_2988|M.vanbaalenii_PYR-1 VSGQLIERIQPARVAALQMQAAIRDQETAVRGYLITADLRFLKPYDEGRA
Mflv_3269|M.gilvum_PYR-GCK LSGQLINNIQPARVAAYQLQAALRDQETAVRGYLIAADQQFLLPYDEGRV
MSMEG_6130|M.smegmatis_MC2_155 LSDELINEIQPARVAAYQLQAALRDQETAVRGYAIAADPQFLAPYDEGQQ
TH_2175|M.thermoresistible__bu MTRELKNDIQPARVAAYQLQSALRDQETAVRSYALTGDEGFLTLYETAQR
MMAR_1264|M.marinum_M VSRELSDNIEPARVAAFQLQSALRDQESGIRGYVISSDSQFLSPYYDGRR
MUL_2624|M.ulcerans_Agy99 VSRELSDNIEPARVAAFQLQSALRDQESGIRGYVISSDSQFLSPYYDGRR
MAV_4241|M.avium_104 VSRQVADGVGPARVAAARLQAALRDQETGLRGYLIAADRQFLAPYYDGQR
Mb0781|M.bovis_AF2122/97 VASGIAVTSMLQHRLTSRIDRVLLEEAQ---IWAQITLPLAPDPYPIHNP
Rv0758|M.tuberculosis_H37Rv VASGIAVTSMLQHRLTSRIDRVLLEEAQ---IWAQITLPLAPDPYPGHNP
MAB_0674|M.abscessus_ATCC_1997 VASGIAVTTTMQRTMMNRADRELIDAAN---GWANR--PPPPPPAFETTR
MLBr_02124|M.leprae_Br4923 LIVGTVVWVGITNDRKERLDRKLDEAAG----FAIPFVPRGLDEIPRSPN
: . : : : : :
Mvan_2988|M.vanbaalenii_PYR-1 AEQQASADIRQLEQDRP---DLLDDLDAIEQASGSWRTNYAEPVVADVRP
Mflv_3269|M.gilvum_PYR-GCK VEQEAAANIRQLEQNRP---DLLDDLDAIEQASGAWRVEYAEPVIADVRP
MSMEG_6130|M.smegmatis_MC2_155 AEADAAASIRDDLDGRD---ELLADLAVIEKAAAAWRAAYADPMIASIQP
TH_2175|M.thermoresistible__bu DERAAADYLRALVGDRP---EQMSDLADIERNAATWRTDYVAPLLSAVRP
MMAR_1264|M.marinum_M TEEAAAQNIRRLLGRRG---DLLADLDAIEKAARAWRASYAEPLIAQVNS
MUL_2624|M.ulcerans_Agy99 TEEAAAQNIRRLLGRRG---DLLADLDAIEKAARAWRASYAEPLIAQVNS
MAV_4241|M.avium_104 TEQAAADEIRRLVGGNAGNTELIADLDAVESAAADWRTNYAEPVIASVTP
Mb0781|M.bovis_AF2122/97 DRPPSRFYVRVISPDGQ----------------SYTALNDNT-AIPAVPA
Rv0758|M.tuberculosis_H37Rv DRPPSRFYVRVISPDGQ----------------SYTALNDNT-AIPAVPA
MAB_0674|M.abscessus_ATCC_1997 HRPPSLFFIRTTDDRGR----------------VVIMLNDRARADPDLPF
MLBr_02124|M.leprae_Br4923 DQDAIITVRRGNLVKSN------------------------------FDI
. * .
Mvan_2988|M.vanbaalenii_PYR-1 GDPGSVDPELTAIGQADFDALRVLFDTQNENLAQARDADIAELERMRTLR
Mflv_3269|M.gilvum_PYR-GCK GTPGPVDPALTEIGKNYSNTLRTLFDTQNEHLAQARNDGVAELERSRTLR
MSMEG_6130|M.smegmatis_MC2_155 GRPHIGNDMVAERGKAQFDQLRGLFDVQNRHLSEARNVGIEDMERMRTWR
TH_2175|M.thermoresistible__bu -TAQPELTALVEGERVEFDRMRALFDVQNSHLTETRQAAVDELERTRGWR
MMAR_1264|M.marinum_M NAPVMLNGAATAIGKAQFDQLRTLFEDQNRHLGAARAQAAERLQRMDDWR
MUL_2624|M.ulcerans_Agy99 NAPVMLNGAATAIGKAQFDQLRTLFEDQNRHLGAARAQAAERLQRMDDWR
MAV_4241|M.avium_104 RAPAVVSAPTADLGKAKFDHLRALFETQNTHLAQAAASATAELHEINGWR
Mb0781|M.bovis_AF2122/97 NNDVGRHPTTLPSIGGSKTLWRAVSVRASDGYLTTVAIDLADVRSTVRSL
Rv0758|M.tuberculosis_H37Rv NNDVGRHPTTLPSIGGSKTLWRAVSVRASDGYLTTVAIDLADVRSTVRSL
MAB_0674|M.abscessus_ATCC_1997 DNDVGGRPVTVRSSDGG-TEWRAVSRHDALGGYTTVARDITDVRTTVRDL
MLBr_02124|M.leprae_Br4923 TLPKLTNDYADTYLRGVRYRVRTVEIPAPEPTSIAVGATYDATVAETNNL
* : :
Mvan_2988|M.vanbaalenii_PYR-1 NAVLLAMAVTFAVTVVALAVLLRRALVRPLSRLAASCRRIAEGNFGERIV
Mflv_3269|M.gilvum_PYR-GCK NAILLAMAVTFGLTIVLLAVVLRRALVRPLDALAAACRRIAEGNFGERIV
MSMEG_6130|M.smegmatis_MC2_155 NSVLIAMIVAFFAMAVVLAVLVRNAVNRPLAALAASCRRITEGNFTERIE
TH_2175|M.thermoresistible__bu NAVLIAMVTAFFVTAALLAVLVRATITRPLEELAAACRRITEGDFGERIS
MMAR_1264|M.marinum_M DRVLAAMVLLFFATAALLGILTRRAVTIPLAALAAACRRVTEGNFGERIV
MUL_2624|M.ulcerans_Agy99 DRVLAAMVLLFFARAALLGVLTRRAVTIPLAALAAACRRFTEGNFGERIV
MAV_4241|M.avium_104 DWVLGAMVLAILGTGLFLGLFSRAAITRPIAQLAASCRRITEGNFAETIV
Mb0781|M.bovis_AF2122/97 VLLQVGIGSAVLVVLGVAGYAVVRRSLRPLAEFEQTAAAIGAGQLDRRVP
Rv0758|M.tuberculosis_H37Rv VLLQVGIGSAVLVVPGVAGYAVVRRSLRPLAEFEQTAAAIGAGQLDRRVP
MAB_0674|M.abscessus_ATCC_1997 LWLQVAIGIAVLTVLGVAGYWLVHRSLRPLAEVEATAADIAAGHLDRRVP
MLBr_02124|M.leprae_Br4923 HRRVLLICGFAIAAAAVFAWLLAAFAVRPFKQLAQQTRSVDAGGEAPRVE
: . *: . . * :
Mvan_2988|M.vanbaalenii_PYR-1 -PQGPRDIRLISLDVEHMRQRIVEELEASR-------AATEALDEQAEEL
Mflv_3269|M.gilvum_PYR-GCK -PHGPRDIRLISLDVEHMRQRIVSELEGSR-------AATDALDVHAEEL
MSMEG_6130|M.smegmatis_MC2_155 -PKGPKDIRAIAIDVEDMRQRIVDELDASK-------QARLALDEQAEEL
TH_2175|M.thermoresistible__bu -LRGPRDLRSIATDVDDMRQRIVDELRTSHRAHAKLSEQTEALDEQAAEL
MMAR_1264|M.marinum_M APRRPRDIRGIAIDVENMRQRMVEELDTSR-------CARAQLDEQTEEL
MUL_2624|M.ulcerans_Agy99 APRRPRDIRGIAIDIENMRQRMVEELDTSR-------CARAQLDEQAEEL
MAV_4241|M.avium_104 APKRPKDIRDMAIDVENMRRRMVEELDVSR-------SAQEQLDEQAAEL
Mb0781|M.bovis_AF2122/97 QWHPRTEVGRLSLALNGMLAQIQRAVASAE--------------SSAEKA
Rv0758|M.tuberculosis_H37Rv QWHPRTEVGRLSLALNGMLAQIQRAVASAE--------------SSAEKA
MAB_0674|M.abscessus_ATCC_1997 ERDPRTEVGRLSLALNGMLAQIQRAVADSE--------------KSAKAA
MLBr_02124|M.leprae_Br4923 -VHGATEAVEIAEAMRGMLQRIWNEQNRTK--------------------
: :: : * :: :.
Mvan_2988|M.vanbaalenii_PYR-1 RRSNAELEQFAYVASHDLQEPLRKVASFCQLLEKRYGDQLDERGVEYIGF
Mflv_3269|M.gilvum_PYR-GCK RRSNAELEQFAYVASHDLQEPLRKVASFCQLLEKRYGDQLDERGKEYIGF
MSMEG_6130|M.smegmatis_MC2_155 RRSNAELEQFAYVASHDLQEPLRKVASFCQLLERRYGDKLDERGVEYIGF
TH_2175|M.thermoresistible__bu RRSNAELEQFAYVASHDLQEPLRKVASFCQLLEKRYGDQLDERGVEYIRF
MMAR_1264|M.marinum_M RRSNAELEQFAYVASHDLQEPLRKVAAFCQLLERRYADQLDERGIEYIGF
MUL_2624|M.ulcerans_Agy99 RRSNAELEQFAYVASHDLQEPLRKVAAFCQLLERRYADQLDERGIEYIGF
MAV_4241|M.avium_104 RRSNTELEQFAYVASHDLQEPLRKVASFCQLLERRYGDQLDERGLEYIGF
Mb0781|M.bovis_AF2122/97 RDSEDRMRQFITDASHELRTPLTTIRGFAELYRQGAARDVG----MLLSR
Rv0758|M.tuberculosis_H37Rv RDSEDRMRQFITDASHELRTPLTTIRGFAELYRQGAARDVG----MLLSR
MAB_0674|M.abscessus_ATCC_1997 RTSEERMRRFITDASHELRTPLTTIRGYAELYRQGAATDTE----LVLGR
MLBr_02124|M.leprae_Br4923 -EALASARDFAAVSSHELRTPLTAMRTNLEVLAT--LDLADDQRKEVLGD
: . * :**:*: ** : :: :
Mvan_2988|M.vanbaalenii_PYR-1 AVDGAKRMQILINDLLTFSRVGRLN-ATETEVDLDEALDNALSNLATAVE
Mflv_3269|M.gilvum_PYR-GCK AVDGAKRMQVLINDLLTFSRVGRLN-ATESDVDLDAALDAALGNLSASVE
MSMEG_6130|M.smegmatis_MC2_155 AVDGAKRMQVLINDLLTFSRVGRLY-STTTEVDLNAVVDAALNNISTAVE
TH_2175|M.thermoresistible__bu AVDGAKRMQVLINDLLTFSRVGRLN-ATEAAVSLDAALNTALDNLSTAIE
MMAR_1264|M.marinum_M AVDGAKRMQVLINDLLSFSRVGRLG-ATSVEVELGATLDAAVANLGAAIE
MUL_2624|M.ulcerans_Agy99 AVDGAKRMQVLINDLLSFSRVGRLG-ATSVEVELGATLDAAVANLGAAIE
MAV_4241|M.avium_104 AVDGAKRMQALINDLLTFSRVGRLG-TTEAEVQLDATLDAGLANLAAAVE
Mb0781|M.bovis_AF2122/97 IESEASRMGLLVDDLLLLARLDAHRPLELCRVDLLALASDAAHDARAMDP
Rv0758|M.tuberculosis_H37Rv IESEASRMGLLVDDLLLLARLDAHRPLELCRVDLLALASDAAHDARAMDP
MAB_0674|M.abscessus_ATCC_1997 IEGESTRMGQLVEDLLLLARLDAHRPMEQRLVDLLVLATDAVHDAKATAP
MLBr_02124|M.leprae_Br4923 VIRTQSRIEATLSALERLAQGELSTSDDHVPVDITELLDRAAHDATRSYP
.*: :. * ::: *.: . :
Mvan_2988|M.vanbaalenii_PYR-1 ESDAVIERPVGG-LPTVKGDPTLLTMLWQNLIGNAVKFRREGVAPRIVID
Mflv_3269|M.gilvum_PYR-GCK ESGAIVDRPTGG-LPTVKGDPTLLTMVWQNLIGNAVKFRREDTAPHIAIE
MSMEG_6130|M.smegmatis_MC2_155 ESGAEIIRPQEP-LPHIDGDPTLLIMVWQNLLGNAVKFRRDGVAPRIVIE
TH_2175|M.thermoresistible__bu ESGTEIVRPAG--LPTVVGDPTLMAMLWQNLIGNAVKFTRPRQRPRIVID
MMAR_1264|M.marinum_M ESDAEIVRLEHD-FPPVVGDPTLLTMLWQNLIGNAVKFRREDLPPRVVID
MUL_2624|M.ulcerans_Agy99 ESDAEIVRLEHG-FPPVVGDPTLLTMLWQNLIGNAVKFRREDLPPRVVID
MAV_4241|M.avium_104 ETGAEIVR-PAE-LPRIVGDPMLLTMLWQNLIGNAIKFRHKDRRPRVVIE
Mb0781|M.bovis_AF2122/97 KRRITLEVLDGPGTPEVLGDESRLRQVLRNLVANAIQHTPESADVTVRVG
Rv0758|M.tuberculosis_H37Rv KRRITLEVLDGPGTPEVLGDESRLRQVLRNLVANAIQHTPESADVTVRVG
MAB_0674|M.abscessus_ATCC_1997 ERDVELEVFDGPGTPEVIGDELRLRQVLGNLVSNALGHT--TAAVRVRVG
MLBr_02124|M.leprae_Br4923 ELKVSLVPSPTC---IIVGLPAGLRLAVDNAVANAVKHGGATRVQLSAVS
: : : * : * :.**: . :
Mvan_2988|M.vanbaalenii_PYR-1 VAQSGDD---------DAQNWSFSLADNGIGIAAEFTDKVFVIFQRLHGR
Mflv_3269|M.gilvum_PYR-GCK CEQIADG---------ESQNWSITVTDNGIGIAPEFADKVFVIFQRLHGR
MSMEG_6130|M.smegmatis_MC2_155 CHAQTGG---------GVDSWLFTVSDNGIGISEEFVDKVFVIFQRLHGR
TH_2175|M.thermoresistible__bu CRRRGAGEQGDDANHGDGDEWLISVSDNGIGIADEFVDKVFVIFQRLHGR
MMAR_1264|M.marinum_M CQLGTGD---------HQGLWLLSVSDNGIGIAQEFADKVFIIFQRLHGR
MUL_2624|M.ulcerans_Agy99 CQLGSGD---------HQGLWLLSVSDNGIGIAQEFADKVFIIFQRLHGR
MAV_4241|M.avium_104 CAPGEGT---------HDGEWLLSVSDNGIGIPEEFSDKVFVIFQRLHGR
Mb0781|M.bovis_AF2122/97 TE---------------GDDAILEVADDGPGMSQEDALRVFERFYRADSS
Rv0758|M.tuberculosis_H37Rv TE---------------GDDAILEVADDGPGMSQEDALRVFERFYRADSS
MAB_0674|M.abscessus_ATCC_1997 TE---------------GDDAVLEVIDRGPGMTKTDAERVFERFYRADSS
MLBr_02124|M.leprae_Br4923 SR----------------AGVEIAVDDNGSGVPEDERQVVFERFSRGSTA
: : * * *:. ** * *
Mvan_2988|M.vanbaalenii_PYR-1 --DAYSGTGIGLALCKKIVEHHGGTIWIDSSYTEGTRFRFTLPMTLDDT-
Mflv_3269|M.gilvum_PYR-GCK --DAYTGTGIGLALCKKIVEHHGGTIWIDTEFSEGTRFRFTLPVTADDT-
MSMEG_6130|M.smegmatis_MC2_155 --DSYSGTGIGLALCKKIIEHHGGNIWIDTSYTGGTRFCFTLPVMPTNKP
TH_2175|M.thermoresistible__bu --DSYTGTGIGLALCKKIVEHHGGTIWIDTGYADGARFCFTLPVTSTET-
MMAR_1264|M.marinum_M --DSYGGTGLGLALCRKIVEHHGGTVWIDSTYTDGTRFQFTLPVAAPATA
MUL_2624|M.ulcerans_Agy99 --DSYGGTGLGLALCRKIVEHHGGTVWIDSSYTEGTRFQFTLPVAAPATA
MAV_4241|M.avium_104 --EVYAGTGVGLALVKKIVEHHGGTVRIDTSYTDGTRFEFSLPVPGP--V
Mb0781|M.bovis_AF2122/97 RARASGGTGLGLSIVDSLVAAHGGAVTVTTALGEGCCFRVSLPRVS----
Rv0758|M.tuberculosis_H37Rv RARASGGTGLGLSIVDSLVAAHGGAVTVTTALGEGCCFRVSLPRVS----
MAB_0674|M.abscessus_ATCC_1997 RTRSSGGTGLGLSIVDALTAAHGGTVTVHTAPGAGCRFEVRLPRADMTDD
MLBr_02124|M.leprae_Br4923 ---SHSGSGLGLALVAQQAQLHGGTASLETSPLGGARLLLRISAPS----
*:*:**:: *** : : * : . :.
Mvan_2988|M.vanbaalenii_PYR-1 ---ATPLLEGSTP-------------------
Mflv_3269|M.gilvum_PYR-GCK ---ADKSLEESPT-------------------
MSMEG_6130|M.smegmatis_MC2_155 GVLTDAQLEGNNT-------------------
TH_2175|M.thermoresistible__bu ---ASAVLEGDQE-------------------
MMAR_1264|M.marinum_M DPEHLEPAEGIAE-------------------
MUL_2624|M.ulcerans_Agy99 DPEHLEPAEGIAE-------------------
MAV_4241|M.avium_104 DETDAVALEGAQQ-------------------
Mb0781|M.bovis_AF2122/97 --DVDQ-LSLTPVVPGPP--------------
Rv0758|M.tuberculosis_H37Rv --DVDQ-LSLTPVVPGPP--------------
MAB_0674|M.abscessus_ATCC_1997 DADVDEPESATPDAAASEPKIVRASRSESAQP
MLBr_02124|M.leprae_Br4923 --------------------------------