For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RLTVQGWQNLVLSVMGVVVFTGAIAGAVLVGRTDTLSDELINEIQPARVAAYQLQAALRDQETAVRGYAI AADPQFLAPYDEGQQAEADAAASIRDDLDGRDELLADLAVIEKAAAAWRAAYADPMIASIQPGRPHIGND MVAERGKAQFDQLRGLFDVQNRHLSEARNVGIEDMERMRTWRNSVLIAMIVAFFAMAVVLAVLVRNAVNR PLAALAASCRRITEGNFTERIEPKGPKDIRAIAIDVEDMRQRIVDELDASKQARLALDEQAEELRRSNAE LEQFAYVASHDLQEPLRKVASFCQLLERRYGDKLDERGVEYIGFAVDGAKRMQVLINDLLTFSRVGRLYS TTTEVDLNAVVDAALNNISTAVEESGAEIIRPQEPLPHIDGDPTLLIMVWQNLLGNAVKFRRDGVAPRIV IECHAQTGGGVDSWLFTVSDNGIGISEEFVDKVFVIFQRLHGRDSYSGTGIGLALCKKIIEHHGGNIWID TSYTGGTRFCFTLPVMPTNKPGVLTDAQLEGNNT*
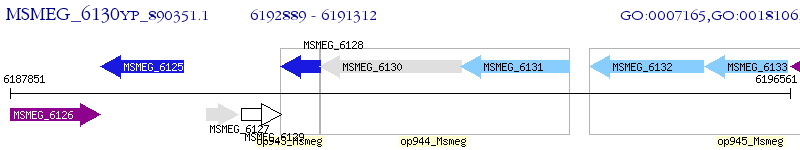
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6130 | - | - | 100% (525) | two-component sensor protein |
| M. smegmatis MC2 155 | MSMEG_2915 | - | 2e-19 | 27.25% (334) | sensor histidine kinase |
| M. smegmatis MC2 155 | MSMEG_5870 | - | 2e-18 | 24.71% (340) | sensor histidine kinase PhoR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1061c | trcS | 1e-19 | 27.58% (330) | two component sensor histidine kinase TRCS |
| M. gilvum PYR-GCK | Mflv_3269 | - | 0.0 | 68.06% (504) | multi-sensor signal transduction histidine kinase |
| M. tuberculosis H37Rv | Rv1032c | trcS | 1e-19 | 27.58% (330) | two component sensor histidine kinase TRCS |
| M. leprae Br4923 | MLBr_02124 | - | 2e-19 | 28.25% (315) | sensor histidine kinase |
| M. abscessus ATCC 19977 | MAB_0674 | - | 2e-17 | 26.39% (341) | putative sensor histidine kinase PhoR |
| M. marinum M | MMAR_1264 | - | 0.0 | 65.94% (505) | two-component system membrane associated sensor kinase |
| M. avium 104 | MAV_4241 | - | 1e-175 | 62.87% (509) | two-component sensor protein |
| M. thermoresistible (build 8) | TH_2175 | - | 0.0 | 62.38% (537) | two-component sensor protein |
| M. ulcerans Agy99 | MUL_2624 | - | 0.0 | 66.14% (505) | two component system membrane associated sensor kinase |
| M. vanbaalenii PYR-1 | Mvan_2988 | - | 0.0 | 68.25% (504) | multi-sensor signal transduction histidine kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3269|M.gilvum_PYR-GCK MSPD---QRGVR---ALTVQGWQYVVLAVMGLVVLVGGIAVSVLLHRTDQ
Mvan_2988|M.vanbaalenii_PYR-1 MSPD---QRGVR---ALTVQGWQYLVLSVMGFVVLTGGIMVAVLLNRTDQ
TH_2175|M.thermoresistible__bu VTPA---ATGERRGSRLTVQGWQNLVLSGMGVIVLAGAITGWLLLHRTDD
MSMEG_6130|M.smegmatis_MC2_155 ----------MR----LTVQGWQNLVLSVMGVVVFTGAIAGAVLVGRTDT
MMAR_1264|M.marinum_M MTPEPPIRPRTRRRIALTVRGWQALVLSIMGVMVLAGTVAGAVLLNRTDD
MUL_2624|M.ulcerans_Agy99 MTPEPPIRPRTRRRIALTVRGWQALVLSIMGVMVLAGTVAGAVLLNRTDG
MAV_4241|M.avium_104 MT-----APRPLRLTQLTVRGWLALVLWIMGTTVLIGALVGAVLLHRTDQ
Mb1061c|M.bovis_AF2122/97 -------MIPDRNTRSRKAPCWRPRSLRQQ---LLLGVLAVVTVVLVAVG
Rv1032c|M.tuberculosis_H37Rv -------MIPDRNTRSRKAPCWRPRSLRQQ---LLLGVLAVVTVVLVAVG
MAB_0674|M.abscessus_ATCC_1997 -------------------MAWDTYCASRDERLREEQLALLVAVPGVRRG
MLBr_02124|M.leprae_Br4923 -------------------------------MNILSRIFARTPSLRTRVV
Mflv_3269|M.gilvum_PYR-GCK LSGQLINNIQPARVAAYQLQAALRDQETAVRGYLIAADQQFLLPYDEGRV
Mvan_2988|M.vanbaalenii_PYR-1 VSGQLIERIQPARVAALQMQAAIRDQETAVRGYLITADLRFLKPYDEGRA
TH_2175|M.thermoresistible__bu MTRELKNDIQPARVAAYQLQSALRDQETAVRSYALTGDEGFLTLYETAQR
MSMEG_6130|M.smegmatis_MC2_155 LSDELINEIQPARVAAYQLQAALRDQETAVRGYAIAADPQFLAPYDEGQQ
MMAR_1264|M.marinum_M VSRELSDNIEPARVAAFQLQSALRDQESGIRGYVISSDSQFLSPYYDGRR
MUL_2624|M.ulcerans_Agy99 VSRELSDNIEPARVAAFQLQSALRDQESGIRGYVISSDSQFLSPYYDGRR
MAV_4241|M.avium_104 VSRQVADGVGPARVAAARLQAALRDQETGLRGYLIAADRQFLAPYYDGQR
Mb1061c|M.bovis_AF2122/97 VVSVLSLSGYVTAMNDAELVESLHALNHSYTRYRDSAQT-------STPT
Rv1032c|M.tuberculosis_H37Rv VVSVLSLSGYVTAMNDAELVESLHALNHSYTRYRDSAQT-------STPT
MAB_0674|M.abscessus_ATCC_1997 VPLRVSLVVGTLVLVALGLVASGIAVTTTMQRTMMNRADRELIDAANGWA
MLBr_02124|M.leprae_Br4923 VATAIGAAIPVLIVGTVVWVGITNDRKERLDRKLDEAAGFAIPFVPRGLD
: : :
Mflv_3269|M.gilvum_PYR-GCK VEQEAAANIRQLEQNRP---DLLDDLDAIEQASGAWRVEYAEPVIADVRP
Mvan_2988|M.vanbaalenii_PYR-1 AEQQASADIRQLEQDRP---DLLDDLDAIEQASGSWRTNYAEPVVADVRP
TH_2175|M.thermoresistible__bu DERAAADYLRALVGDRP---EQMSDLADIERNAATWRTDYVAPLLSAVRP
MSMEG_6130|M.smegmatis_MC2_155 AEADAAASIRDDLDGRD---ELLADLAVIEKAAAAWRAAYADPMIASIQP
MMAR_1264|M.marinum_M TEEAAAQNIRRLLGRRG---DLLADLDAIEKAARAWRASYAEPLIAQVNS
MUL_2624|M.ulcerans_Agy99 TEEAAAQNIRRLLGRRG---DLLADLDAIEKAARAWRASYAEPLIAQVNS
MAV_4241|M.avium_104 TEQAAADEIRRLVGGNAGNTELIADLDAVESAAADWRTNYAEPVIASVTP
Mb1061c|M.bovis_AF2122/97 GNLPMSQAVLEFTGQTPG-----NLIAVLHDGVVIGSAVFSEDGARPAPP
Rv1032c|M.tuberculosis_H37Rv GNLPMSQAVLEFTGQTPG-----NLIAVLHDGVVIGSAVFSEDGARPAPP
MAB_0674|M.abscessus_ATCC_1997 NRPPPPPPAFETTRHRP------PSLFFIRTTDDRGRVVIMLNDRARADP
MLBr_02124|M.leprae_Br4923 EIPRSPNDQDAIITVRRG--------NLVKSNFDITLPKLTNDYADTYLR
. :.
Mflv_3269|M.gilvum_PYR-GCK GTPGPVDPALTEIGKNYSNTLRTLFDTQNEHLAQARN-----DGVAELER
Mvan_2988|M.vanbaalenii_PYR-1 GDPGSVDPELTAIGQADFDALRVLFDTQNENLAQARD-----ADIAELER
TH_2175|M.thermoresistible__bu -TAQPELTALVEGERVEFDRMRALFDVQNSHLTETRQ-----AAVDELER
MSMEG_6130|M.smegmatis_MC2_155 GRPHIGNDMVAERGKAQFDQLRGLFDVQNRHLSEARN-----VGIEDMER
MMAR_1264|M.marinum_M NAPVMLNGAATAIGKAQFDQLRTLFEDQNRHLGAARA-----QAAERLQR
MUL_2624|M.ulcerans_Agy99 NAPVMLNGAATAIGKAQFDQLRTLFEDQNRHLGAARA-----QAAERLQR
MAV_4241|M.avium_104 RAPAVVSAPTADLGKAKFDHLRALFETQNTHLAQAAA-----SATAELHE
Mb1061c|M.bovis_AF2122/97 DVIRAIEAQVWDGGPPRVESLGSLGAYQVDSSAAGADRLFVGVSLSLANQ
Rv1032c|M.tuberculosis_H37Rv DVIRAIEAQVWDGGPPRVESLGSLGAYQVDSSAAGADRLFVGVSLSLANQ
MAB_0674|M.abscessus_ATCC_1997 DLPFDNDVGGRPVTVRSSDGGTEWRAVSRHDALGGYTT--VARDITDVRT
MLBr_02124|M.leprae_Br4923 GVRYRVRTVEIPAPEPTSIAVGATYDATVAETNNLHR-------------
Mflv_3269|M.gilvum_PYR-GCK SRTLRNAILLAMAVTFGLTIVLLAVVLRRALVRPLDALAAACRRIAEGNF
Mvan_2988|M.vanbaalenii_PYR-1 MRTLRNAVLLAMAVTFAVTVVALAVLLRRALVRPLSRLAASCRRIAEGNF
TH_2175|M.thermoresistible__bu TRGWRNAVLIAMVTAFFVTAALLAVLVRATITRPLEELAAACRRITEGDF
MSMEG_6130|M.smegmatis_MC2_155 MRTWRNSVLIAMIVAFFAMAVVLAVLVRNAVNRPLAALAASCRRITEGNF
MMAR_1264|M.marinum_M MDDWRDRVLAAMVLLFFATAALLGILTRRAVTIPLAALAAACRRVTEGNF
MUL_2624|M.ulcerans_Agy99 MDDWRDRVLAAMVLLFFARAALLGVLTRRAVTIPLAALAAACRRFTEGNF
MAV_4241|M.avium_104 INGWRDWVLGAMVLAILGTGLFLGLFSRAAITRPIAQLAASCRRITEGNF
Mb1061c|M.bovis_AF2122/97 IIARKKVTTVALVGAALVVTAALTVWVVGYALRPLRRVAATAAEVAT---
Rv1032c|M.tuberculosis_H37Rv IIARKKVTTVALVGAALVVTAALTVWVVGYALRPLRRVAATAAEVAT---
MAB_0674|M.abscessus_ATCC_1997 TVRDLLWLQVAIGIAVLTVLGVAGYWLVHRSLRPLAEVEATAADIAAGHL
MLBr_02124|M.leprae_Br4923 -------RVLLICGFAIAAAAVFAWLLAAFAVRPFKQLAQQTRSVDAGG-
: *: : .
Mflv_3269|M.gilvum_PYR-GCK GERIV-PHGPRDIRLISLDVEHMRQRIVSELEGSR-------AATDALDV
Mvan_2988|M.vanbaalenii_PYR-1 GERIV-PQGPRDIRLISLDVEHMRQRIVEELEASR-------AATEALDE
TH_2175|M.thermoresistible__bu GERIS-LRGPRDLRSIATDVDDMRQRIVDELRTSHRAHAKLSEQTEALDE
MSMEG_6130|M.smegmatis_MC2_155 TERIE-PKGPKDIRAIAIDVEDMRQRIVDELDASK-------QARLALDE
MMAR_1264|M.marinum_M GERIVAPRRPRDIRGIAIDVENMRQRMVEELDTSR-------CARAQLDE
MUL_2624|M.ulcerans_Agy99 GERIVAPRRPRDIRGIAIDIENMRQRMVEELDTSR-------CARAQLDE
MAV_4241|M.avium_104 AETIVAPKRPKDIRDMAIDVENMRRRMVEELDVSR-------SAQEQLDE
Mb1061c|M.bovis_AF2122/97 -MPLTDDDHQISVRVRPGDTDPDNEVGIVGHTLNR--------LLDNVDG
Rv1032c|M.tuberculosis_H37Rv -MPLTDDDHQISVRVRPGDTDPDNEVGIVGHTLNR--------LLDNVDG
MAB_0674|M.abscessus_ATCC_1997 DRRVPERDPRTEVGRLSLALNGMLAQIQRAVADSE--------------K
MLBr_02124|M.leprae_Br4923 EAPRVEVHGATEAVEIAEAMRGMLQRIWNEQNRTK---------------
. . ..
Mflv_3269|M.gilvum_PYR-GCK HAEELRRSNAELEQFAYVASHDLQEPLRKVASFCQLLEKRYGDQLDERGK
Mvan_2988|M.vanbaalenii_PYR-1 QAEELRRSNAELEQFAYVASHDLQEPLRKVASFCQLLEKRYGDQLDERGV
TH_2175|M.thermoresistible__bu QAAELRRSNAELEQFAYVASHDLQEPLRKVASFCQLLEKRYGDQLDERGV
MSMEG_6130|M.smegmatis_MC2_155 QAEELRRSNAELEQFAYVASHDLQEPLRKVASFCQLLERRYGDKLDERGV
MMAR_1264|M.marinum_M QTEELRRSNAELEQFAYVASHDLQEPLRKVAAFCQLLERRYADQLDERGI
MUL_2624|M.ulcerans_Agy99 QAEELRRSNAELEQFAYVASHDLQEPLRKVAAFCQLLERRYADQLDERGI
MAV_4241|M.avium_104 QAAELRRSNTELEQFAYVASHDLQEPLRKVASFCQLLERRYGDQLDERGL
Mb1061c|M.bovis_AF2122/97 ALAHRVDSDLRMRQFITDASHELRTPLAAIQGYAELTRQDSSDLPPTTEY
Rv1032c|M.tuberculosis_H37Rv ALAHRVDSDLRMRQFITDASHELRTPLAAIQGYAELTRQDSSDLPPTTEY
MAB_0674|M.abscessus_ATCC_1997 SAKAARTSEERMRRFITDASHELRTPLTTIRGYAELYRQG---AATDTEL
MLBr_02124|M.leprae_Br4923 ------EALASARDFAAVSSHELRTPLTAMR--TNLEVLATLDLADDQRK
: . * :**:*: ** : :*
Mflv_3269|M.gilvum_PYR-GCK EYIGFAVDGAKRMQVLINDLLTFSRVGRLNATESDVDLDAALDAALGNLS
Mvan_2988|M.vanbaalenii_PYR-1 EYIGFAVDGAKRMQILINDLLTFSRVGRLNATETEVDLDEALDNALSNLA
TH_2175|M.thermoresistible__bu EYIRFAVDGAKRMQVLINDLLTFSRVGRLNATEAAVSLDAALNTALDNLS
MSMEG_6130|M.smegmatis_MC2_155 EYIGFAVDGAKRMQVLINDLLTFSRVGRLYSTTTEVDLNAVVDAALNNIS
MMAR_1264|M.marinum_M EYIGFAVDGAKRMQVLINDLLSFSRVGRLGATSVEVELGATLDAAVANLG
MUL_2624|M.ulcerans_Agy99 EYIGFAVDGAKRMQVLINDLLSFSRVGRLGATSVEVELGATLDAAVANLG
MAV_4241|M.avium_104 EYIGFAVDGAKRMQALINDLLTFSRVGRLGTTEAEVQLDATLDAGLANLA
Mb1061c|M.bovis_AF2122/97 ALARIESE-ARRMTLLVDELLLLSRLSEGEDLETEDLDLTDLVINAVNDA
Rv1032c|M.tuberculosis_H37Rv ALARIESE-ARRMTLLVDELLLLSRLSEGEDLETEDLDLTDLVINAVNDA
MAB_0674|M.abscessus_ATCC_1997 VLGRIEGE-STRMGQLVEDLLLLARLDAHRPMEQRLVDLLVLATDAVHDA
MLBr_02124|M.leprae_Br4923 EVLGDVIRTQSRIEATLSALERLAQGELSTSDDHVPVDITELLDRAAHDA
*: :. * ::: : : .
Mflv_3269|M.gilvum_PYR-GCK ASVEESGAIVDRPTGG--LPTVKGDPTLLTMVWQNLIGNAVKFRREDTAP
Mvan_2988|M.vanbaalenii_PYR-1 TAVEESDAVIERPVGG--LPTVKGDPTLLTMLWQNLIGNAVKFRREGVAP
TH_2175|M.thermoresistible__bu TAIEESGTEIVRPAG---LPTVVGDPTLMAMLWQNLIGNAVKFTRPRQRP
MSMEG_6130|M.smegmatis_MC2_155 TAVEESGAEIIRPQEP--LPHIDGDPTLLIMVWQNLLGNAVKFRRDGVAP
MMAR_1264|M.marinum_M AAIEESDAEIVRLEHD--FPPVVGDPTLLTMLWQNLIGNAVKFRREDLPP
MUL_2624|M.ulcerans_Agy99 AAIEESDAEIVRLEHG--FPPVVGDPTLLTMLWQNLIGNAVKFRREDLPP
MAV_4241|M.avium_104 AAVEETGAEIVR-PAE--LPRIVGDPMLLTMLWQNLIGNAIKFRHKDRRP
Mb1061c|M.bovis_AF2122/97 AVAAPTHRWVKNLPDE--PVWVNGDHARLHQLVSNLLTNAWVHTQPGVTV
Rv1032c|M.tuberculosis_H37Rv AVAAPTHRWVKNLPDE--PVWVNGDHARLHQLVSNLLTNAWVHTQPGVTV
MAB_0674|M.abscessus_ATCC_1997 KATAPERDVELEVFDGPGTPEVIGDELRLRQVLGNLVSNALGHTTAAVRV
MLBr_02124|M.leprae_Br4923 TRSYPELKVSLVPSPT---CIIVGLPAGLRLAVDNAVANAVKHGG---AT
: * : * : ** .
Mflv_3269|M.gilvum_PYR-GCK HIAIECEQIADG---------ESQNWSITVTDNGIGIAPEFADKVFVIFQ
Mvan_2988|M.vanbaalenii_PYR-1 RIVIDVAQSGDD---------DAQNWSFSLADNGIGIAAEFTDKVFVIFQ
TH_2175|M.thermoresistible__bu RIVIDCRRRGAGEQGDDANHGDGDEWLISVSDNGIGIADEFVDKVFVIFQ
MSMEG_6130|M.smegmatis_MC2_155 RIVIECHAQTGG---------GVDSWLFTVSDNGIGISEEFVDKVFVIFQ
MMAR_1264|M.marinum_M RVVIDCQLGTGD---------HQGLWLLSVSDNGIGIAQEFADKVFIIFQ
MUL_2624|M.ulcerans_Agy99 RVVIDCQLGSGD---------HQGLWLLSVSDNGIGIAQEFADKVFIIFQ
MAV_4241|M.avium_104 RVVIECAPGEGT---------HDGEWLLSVSDNGIGIPEEFSDKVFVIFQ
Mb1061c|M.bovis_AF2122/97 TIGITCHRTGPN----------APCVELSVTDDGPDIDPEILPHLFDRFV
Rv1032c|M.tuberculosis_H37Rv TIGITCHRTGPN----------APCVELSVTDDGPDIDPEILPHLFDRFV
MAB_0674|M.abscessus_ATCC_1997 RVGTEG-----------------DDAVLEVIDRGPGMTKTDAERVFERFY
MLBr_02124|M.leprae_Br4923 RVQLSAVSSRAG-------------VEIAVDDNGSGVPEDERQVVFERFS
: : : * * .: :* *
Mflv_3269|M.gilvum_PYR-GCK RLHG-RDAYTG-TGIGLALCKKIVEHHGGTIWIDTEFSEGTRFRFTLPVT
Mvan_2988|M.vanbaalenii_PYR-1 RLHG-RDAYSG-TGIGLALCKKIVEHHGGTIWIDSSYTEGTRFRFTLPMT
TH_2175|M.thermoresistible__bu RLHG-RDSYTG-TGIGLALCKKIVEHHGGTIWIDTGYADGARFCFTLPVT
MSMEG_6130|M.smegmatis_MC2_155 RLHG-RDSYSG-TGIGLALCKKIIEHHGGNIWIDTSYTGGTRFCFTLPVM
MMAR_1264|M.marinum_M RLHG-RDSYGG-TGLGLALCRKIVEHHGGTVWIDSTYTDGTRFQFTLPVA
MUL_2624|M.ulcerans_Agy99 RLHG-RDSYGG-TGLGLALCRKIVEHHGGTVWIDSSYTEGTRFQFTLPVA
MAV_4241|M.avium_104 RLHG-REVYAG-TGVGLALVKKIVEHHGGTVRIDTSYTDGTRFEFSLPVP
Mb1061c|M.bovis_AF2122/97 RASKSRSNGSG-HGLGLAIVSSIVKAHRGSVTAESGNGQ-TVFRVRLPMI
Rv1032c|M.tuberculosis_H37Rv RASKSRSNGSG-HGLGLAIVSSIVKAHRGSVTAESGNGQ-TVFRVRLPMI
MAB_0674|M.abscessus_ATCC_1997 RADSSRTRSSGGTGLGLSIVDALTAAHGGTVTVHTAPGAGCRFEVRLPRA
MLBr_02124|M.leprae_Br4923 RGST--ASHSG-SGLGLALVAQQAQLHGGTASLETSPLGGARLLLRISAP
* * *:**:: * *. .: : . :.
Mflv_3269|M.gilvum_PYR-GCK ADDT----ADKSLEESPT-------------------
Mvan_2988|M.vanbaalenii_PYR-1 LDDT----ATPLLEGSTP-------------------
TH_2175|M.thermoresistible__bu STET----ASAVLEGDQE-------------------
MSMEG_6130|M.smegmatis_MC2_155 PTNKPGVLTDAQLEGNNT-------------------
MMAR_1264|M.marinum_M APATADPEHLEPAEGIAE-------------------
MUL_2624|M.ulcerans_Agy99 APATADPEHLEPAEGIAE-------------------
MAV_4241|M.avium_104 GP--VDETDAVALEGAQQ-------------------
Mb1061c|M.bovis_AF2122/97 EQQIATTA-----------------------------
Rv1032c|M.tuberculosis_H37Rv EQQIATTA-----------------------------
MAB_0674|M.abscessus_ATCC_1997 DMTDDDADVDEPESATPDAAASEPKIVRASRSESAQP
MLBr_02124|M.leprae_Br4923 S------------------------------------