For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TAILIAEDEPRVSSFVRKGLSSNGYSVTLVTDGRSAYFSARNGGFDLMLLDTDLPGMDGFAVLRRLRADG SPLPVVMLMSRAGVADTTAALEEGADDYVVKPFRFEDLLARVRLRLTAGRAASTTLAYGSLLLDLRTRRA HVGDYAVDLSARECKLAETFLRHPGQVLSREQLLNQVWGRDHETGSNVVDVYVRYLRRKLGANRFATLRG MGYRLEPI*
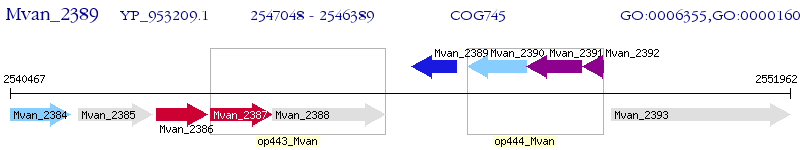
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_2389 | - | - | 100% (219) | two component transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_4844 | - | 2e-39 | 40.99% (222) | two component transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_5009 | - | 1e-38 | 41.63% (221) | two component transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0618c | tcrA | 7e-42 | 42.20% (218) | putative two component DNA binding transcriptional |
| M. gilvum PYR-GCK | Mflv_3998 | - | 1e-85 | 72.73% (220) | two component transcriptional regulator |
| M. tuberculosis H37Rv | Rv0602c | tcrA | 6e-42 | 42.20% (218) | two component DNA binding transcriptional regulatory protein |
| M. leprae Br4923 | MLBr_02123 | - | 4e-39 | 41.18% (221) | two-component response regulator |
| M. abscessus ATCC 19977 | MAB_1076 | - | 4e-41 | 42.53% (221) | two component response transcriptional regulatory protein MprA |
| M. marinum M | MMAR_4633 | prrA | 2e-38 | 41.18% (221) | two-component response transcriptional regulatory protein |
| M. avium 104 | MAV_1022 | - | 2e-38 | 41.18% (221) | two component response transcriptional regulatory protein prra |
| M. smegmatis MC2 155 | MSMEG_5662 | prrA | 9e-39 | 42.08% (221) | DNA-binding response regulator PrrA |
| M. thermoresistible (build 8) | TH_4055 | - | 2e-81 | 68.18% (220) | PUTATIVE two component transcriptional regulator, winged helix |
| M. ulcerans Agy99 | MUL_0248 | prrA | 5e-38 | 40.72% (221) | two component response transcriptional regulatory protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_2389|M.vanbaalenii_PYR-1 ---------------------MTAILIAEDEPRVSSFVRKGLSSNGYSVT
Mflv_3998|M.gilvum_PYR-GCK ---------------------MAAVLIAEDEPRISALVRKGLSANGFSVT
TH_4055|M.thermoresistible__bu ---------------------MARILIVEDEPRISSFIEKGLSANGFTAT
MMAR_4633|M.marinum_M ------------MGGMDTGANSPRVLVVDDDSDVLASLERGLRLSGFEVS
MUL_0248|M.ulcerans_Agy99 ------------MGGMDTGANSPRVLVVDDDSDVLASLERGLRLSGFEVS
MAV_1022|M.avium_104 ---------------MDTAASSPRVLVVDDDSDVLASLERGLRLSGFEVS
MLBr_02123|M.leprae_Br4923 ---------------MDTGVSSPRVLVVDDDSDVLASLERGLRLSGFEVS
MSMEG_5662|M.smegmatis_MC2_155 ------------MDSTTSGMASPRVLVVDDDPDVLASLERGLRLSGFDVS
MAB_1076|M.abscessus_ATCC_1997 --------------------MPVRILVVDDDRAVRESLRRSLSFNGYSVD
Mb0618c|M.bovis_AF2122/97 MADETTMRAGRGPGRACGRVSGVRILVIEDEPKMTALLARALTEEGHTVD
Rv0602c|M.tuberculosis_H37Rv MADETTMRAGRGPGRACGRVSGVRILVVEDEPKMTALLARALTEEGHTVD
:*: :*: : : :.* .*. .
Mvan_2389|M.vanbaalenii_PYR-1 LVTDGRSAYFSARNGGFDLMLLDTDLPGMDGFAVLRRLRADGSPLPVVML
Mflv_3998|M.gilvum_PYR-GCK VVPDGPSAYAYAHSGAFDLMVLDTGLPGMDGLSVLRRLRAEGSALPVIVL
TH_4055|M.thermoresistible__bu TVTDGPTGYDYAMTGGFDLMVLDIGLPSMDGFTVLRKLREDGNQIPVIVL
MMAR_4633|M.marinum_M TAVDGAEALRSATETRPDAIVLDINMPVLDGVSVVTALRAMDNDVPVCVL
MUL_0248|M.ulcerans_Agy99 TAVDGAEALRSATETRPDAIVLDINMPVLDGVSVVTALRAMDNDVPVCVL
MAV_1022|M.avium_104 TAVDGAEALRSATETRPDAIVLDINMPVLDGVSVVTALRAMDNDVPVCVL
MLBr_02123|M.leprae_Br4923 TAIDGAEALRNATETRPDAIVLDINMPVLDGVSVVTALRAMDNDVPVCVL
MSMEG_5662|M.smegmatis_MC2_155 TAVDGAEALRSATETRPDAIVLDINMPVLDGVSVVTALRAMDNDVPVCVL
MAB_1076|M.abscessus_ATCC_1997 LAEDGVDALNVIANDRPDALVLDVMMPKLDGLEVCRRLRSTGDDLPILVL
Mb0618c|M.bovis_AF2122/97 TVADGRHAVAAVDGGDYDAVVLDVMLPGIDGFEVCARLRRQRVWTPVLML
Rv0602c|M.tuberculosis_H37Rv TVADGRHAVAAVDGGDYDAVVLDVMLPGIDGFEVCARLRRQRVWTPVLML
. ** . * ::** :* :**. * ** *: :*
Mvan_2389|M.vanbaalenii_PYR-1 MSRAGVADTTAALEEGADDYVVKPFRFEDLLARVRLRLT---AGRAAST-
Mflv_3998|M.gilvum_PYR-GCK TTRTTGADT-AALLGGADDVIAKPFRLEDLVSRVRTRLS---PGRAARTH
TH_4055|M.thermoresistible__bu TARDSVEDTVAGLEGGADDYMPKPFRFEELLARIRLRLT---TERSAELT
MMAR_4633|M.marinum_M SARSSVDDRVAGLEAGADDYLVKPFVLAELVARVKALLRRRGATATSSSE
MUL_0248|M.ulcerans_Agy99 SARSSVDDRVAGLEAGADDYLVKPFVLAELVARVKALLRRRGATATSSSE
MAV_1022|M.avium_104 SARSSVDDRVAGLEAGADDYLVKPFVLAELVARVKALLRRRGATATSSSE
MLBr_02123|M.leprae_Br4923 SARSSVDDRVAGLEAGADDYLVKPFVLAELVARVKALLRRRGATATSSSE
MSMEG_5662|M.smegmatis_MC2_155 SARSSVDDRVAGLEAGADDYLVKPFVLAELVARVKALLRRRGSSATFSSE
MAB_1076|M.abscessus_ATCC_1997 TARDSVSERVAGLDAGADDYLPKPFALEELLARMRALLRRTTIDESADAA
Mb0618c|M.bovis_AF2122/97 TARGAVTDRIAGLDGGADDYLTKPFNLDELFARLRALSR---RGPIPRPP
Rv0602c|M.tuberculosis_H37Rv TARGAVTDRIAGLDGGADDYLTKPFNLDELFARLRALSR---RGPIPRPP
:* : *.* **** : *** : :*.:*::
Mvan_2389|M.vanbaalenii_PYR-1 TLAYGSLLLDLRTRRAHVGDYAVDLSARECKLAETFLRHPGQVLSREQLL
Mflv_3998|M.gilvum_PYR-GCK TLSYGNLRLDLRTRRAHVGDYSVDLSAREFALAEMFLRHPGQVLTRDQLL
TH_4055|M.thermoresistible__bu VLTCGGMQLDLRTRRAKVGDKTVDLSAREFALAETFMRHPGQVLSREQLL
MMAR_4633|M.marinum_M TITVGPLEVDIPGRRARVNGVDVDLTKREFDLLAVLAEHKTAVLSRAQLL
MUL_0248|M.ulcerans_Agy99 IITVGPLEVDIPGRRARVNGVDVDLTKREFDLLAVLAEHKTAVLSRAQLL
MAV_1022|M.avium_104 TITVGPLEVDIPGRRARVNGVDVDLTKREFDLLAVLAEHKTAVLSRAQLL
MLBr_02123|M.leprae_Br4923 TIAVGPLEVDIPGRRARVNGVDVDLTKREFDLLAVLAEHKTTVLSRAQLL
MSMEG_5662|M.smegmatis_MC2_155 TIQVGPLEVDIPGRRARVNGVDVDLTKREFDLLAVLAEHKTAVLSRAQLL
MAB_1076|M.abscessus_ATCC_1997 TLSFGDLTLDPVTREVHRGTRSISLTRTEFALLEMLMANPRRVLTRSRIL
Mb0618c|M.bovis_AF2122/97 TLEAGDLRLDPSEHRVWRADTEIRLSHKEFTLLEALIRRPGIVHTRAQLL
Rv0602c|M.tuberculosis_H37Rv TLEAGDLRLDPSEHRVWRADTEIRLSHKEFTLLEALIRRPGIVHTRAQLL
: * : :* :.. : *: * * : . * :* ::*
Mvan_2389|M.vanbaalenii_PYR-1 NQVWGRDHETGSNVVDVYVRYLRRKLGANR----FATLRGMGYRLEPI--
Mflv_3998|M.gilvum_PYR-GCK RQVWGNDYEPGSNVVDVYVRYLRRKLGARR----FVTLRGMGYRLEAVH-
TH_4055|M.thermoresistible__bu SRVWGYDFDPGSNVVDVYVRYLRRKLGAER----FVTVRGMGYRLEELP-
MMAR_4633|M.marinum_M ELVWGYDFAADTNVVDVFIGYLRRKLEANGGPRLLHTVRGVGFVLRMQ--
MUL_0248|M.ulcerans_Agy99 ELVWGYDFAADTNVVDVFIGYLRRKLEANGGPRLLHTVRGVGFVLRMH--
MAV_1022|M.avium_104 ELVWGYDFAADTNVVDVFIGYLRRKLEANGGPRLLHTVRGVGFVLRMQ--
MLBr_02123|M.leprae_Br4923 ELVWGYDFAADTNVVDVFIGYLRRKLEANSGPRLLHTVRGVGFVLRMQ--
MSMEG_5662|M.smegmatis_MC2_155 ELVWGYDFAADTNVVDVFIGYLRRKLEAGGAPRLLHTVRGVGFVLRTQ--
MAB_1076|M.abscessus_ATCC_1997 EEVWGFDFPTSGNALEVYVGYLRRKTEAEGELRLIHTVRGVGYVLRETPP
Mb0618c|M.bovis_AF2122/97 ERCWDAAYEARSNIVDVYIRYLRDKIDRPFGVTSLETIRGAGYRLRKDGG
Rv0602c|M.tuberculosis_H37Rv ERCWDAAYEARSNIVDVYIRYLRDKIDRPFGVTSLETIRGAGYRLRKDGG
*. . . * ::*:: *** * : *:** *: *.
Mvan_2389|M.vanbaalenii_PYR-1 ------
Mflv_3998|M.gilvum_PYR-GCK ------
TH_4055|M.thermoresistible__bu ------
MMAR_4633|M.marinum_M ------
MUL_0248|M.ulcerans_Agy99 ------
MAV_1022|M.avium_104 ------
MLBr_02123|M.leprae_Br4923 ------
MSMEG_5662|M.smegmatis_MC2_155 ------
MAB_1076|M.abscessus_ATCC_1997 ------
Mb0618c|M.bovis_AF2122/97 RHALPR
Rv0602c|M.tuberculosis_H37Rv RHALPR