For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TPPPVARILLADDHALVRSGLRMILDAEPDLRVVVEAADGYQALNALEQTPVDIAILDIAMPRMTGLQAA REISRKYPHVRILILSMHDNEQYFFEALRAGASGYVLKSVADLDLLKACRATLRGEPFVYPGAVTALIQD YLNRARHGDDLPGTIVTSREEEVLKLIAEGYSTREIAGTLGISAKTVDRHRANLLAKLGLRDRLALTRYA IRVGLIEP*
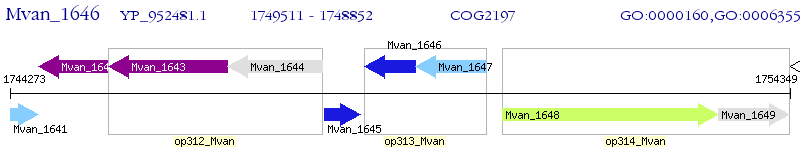
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1646 | - | - | 100% (219) | two component LuxR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_5522 | - | 2e-30 | 37.50% (216) | two component LuxR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_1358 | - | 4e-30 | 35.00% (220) | two component LuxR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3157c | devR | 3e-29 | 32.24% (214) | two component transcriptional regulatory protein DevR |
| M. gilvum PYR-GCK | Mflv_5014 | - | 8e-30 | 34.70% (219) | two component LuxR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3133c | devR | 3e-29 | 32.24% (214) | two component transcriptional regulatory protein DevR |
| M. leprae Br4923 | MLBr_01286 | - | 6e-12 | 36.61% (112) | putative two-component response regulatory protein |
| M. abscessus ATCC 19977 | MAB_1522 | - | 1e-28 | 35.55% (211) | putative two-component system response regulator LuxR |
| M. marinum M | MMAR_4782 | narL | 7e-30 | 34.86% (218) | nitrate/nitrite response regulator protein NarL |
| M. avium 104 | MAV_5280 | - | 2e-32 | 36.87% (217) | two-component system response regulator |
| M. smegmatis MC2 155 | MSMEG_0856 | - | 1e-95 | 82.63% (213) | DNA-binding response regulator, LuxR family protein |
| M. thermoresistible (build 8) | TH_3184 | - | 6e-31 | 36.07% (219) | PUTATIVE two component transcriptional regulator, LuxR family |
| M. ulcerans Agy99 | MUL_0351 | narL | 2e-30 | 35.32% (218) | nitrate/nitrite response regulator protein NarL |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3157c|M.bovis_AF2122/97 ------------MVKVFLVDDHEVVRRGLVDLLGADPE-LDVVGEAGSVA
Rv3133c|M.tuberculosis_H37Rv ------------MVKVFLVDDHEVVRRGLVDLLGADPE-LDVVGEAGSVA
Mvan_1646|M.vanbaalenii_PYR-1 -------MTPPPVARILLADDHALVRSGLRMILDAEPD-LRVVVEAADGY
MSMEG_0856|M.smegmatis_MC2_155 -----------MAARILLADDHALVRSGLRMIIDAEPD-LQVVAEAADGY
MMAR_4782|M.marinum_M -----MDVTKTEKVRVVVGDDHPLFREGVVRALSLSGS-VNVVGEADDGA
MUL_0351|M.ulcerans_Agy99 -----MDVTKTEKVRVVVGDDHPLFREGVVRALSLSGS-VNVVGEADDGA
Mflv_5014|M.gilvum_PYR-GCK --------MSSEPVRIVLVDDHEMVIEGLKAMLAAFADRVCVVGQSVGAD
TH_3184|M.thermoresistible__bu ----MEPVAAPSPVRIVLVDDHEMVIEGLKAMLAAFEDRVCVVGHAVGAE
MAB_1522|M.abscessus_ATCC_1997 ------MTD--TEVGVLLVDDQELVRTGLRRILRRKDG-FAIVGECADGS
MAV_5280|M.avium_104 ------MTDPAPEITVLLVDDQDLVRSGLRRILRRKDG-FVIVAECADGD
MLBr_01286|M.leprae_Br4923 MTGPTTDADAATPHRVLIAEDEALIRLDLAEMLR--DEGYDIVGEAADGQ
:.: :*. :. .: : :* .. .
Mb3157c|M.bovis_AF2122/97 EAMARVPAARPDVAVLDVRLPDGNGIELCRDLLSRMPDLRCLILTSYTSD
Rv3133c|M.tuberculosis_H37Rv EAMARVPAARPDVAVLDVRLPDGNGIELCRDLLSRMPDLRCLILTSYTSD
Mvan_1646|M.vanbaalenii_PYR-1 QALNALEQTPVDIAILDIAMPRMTGLQAAREISRKYPHVRILILSMHDNE
MSMEG_0856|M.smegmatis_MC2_155 EALSALDTETVDLAILDIAMPRMTGLQAAREINRSHPDVRILILSMYDNE
MMAR_4782|M.marinum_M AALELIKTHLPDVALLDYRMPRMDGAQVAAAVRSNELPTRVLLISAHDES
MUL_0351|M.ulcerans_Agy99 AALELIETHLPDVALLDYRMPRMDGAQVAAAVRSNGLPTRVLLISAHDES
Mflv_5014|M.gilvum_PYR-GCK QVMGVVTDLDPDIVLCDVRMQGSSGLDVCQQLRRRDPDRKIVMLSVYDDE
TH_3184|M.thermoresistible__bu RAVEVVAELDPDIVLCDVRMQNTSGLDLCLTLREQAPHRKVVMLSVYDDE
MAB_1522|M.abscessus_ATCC_1997 EVAAAVSRARPDVVIMDLRMKQMSGIEAIRLLHAAQ-GPPALALTTFDDD
MAV_5280|M.avium_104 EVPAAVAAHRPDVVVMDLRMRRVDGIEATRRLGGR---PPVLALTTFNED
MLBr_01286|M.leprae_Br4923 EAVELAERHKPDLVIMDVKMPRRDGIDAASEIASKR-IAPIVVLTAFSQR
. *:.: * : * : : : :: . .
Mb3157c|M.bovis_AF2122/97 EAMLDAILAGASGYVVKDIKGMELARAVKDVGAGRSLLDNRAAAALMAKL
Rv3133c|M.tuberculosis_H37Rv EAMLDAILAGASGYVVKDIKGMELARAVKDVGAGRSLLDNRAAAALMAKL
Mvan_1646|M.vanbaalenii_PYR-1 QYFFEALRAGASGYVLKSVADLDLLKACRATLRGEPFVYPGAVTALIQDY
MSMEG_0856|M.smegmatis_MC2_155 QYFFEALKAGASGYVLKSVADRDLLEACRATLRGEPFLYAGAITALIRDF
MMAR_4782|M.marinum_M AIVYQALQQGAAGFLLKDSTRTEIVKAVLDCAKGRDVVAP----SLVGGL
MUL_0351|M.ulcerans_Agy99 AIVYQALQQGAAGFLLKDSTRTEIVKAVLDCAKGRDVVAP----SLVGGL
Mflv_5014|M.gilvum_PYR-GCK QYLFQAMRVGASGYLLKSISSEDLVEQLESVHSGKTAIDPTMAARAVE--
TH_3184|M.thermoresistible__bu QYLYQALRVGASGYLLKSISSDELVRQLEMVHRGQTALDPGMAARAVDTA
MAB_1522|M.abscessus_ATCC_1997 ELLSGALRAGAAGFILKDSPAEELVRAVHAVARGEAYLDPAVTSRVLNAY
MAV_5280|M.avium_104 ELLSGALRAGAAGFVLKDSSAEELIRAVRAVARGEGYLDPAVTSRVLTTY
MLBr_01286|M.leprae_Br4923 DLVERARDAGAMAYLVKPFTISDLIPAIALAMS--RFSELTALEREVATL
. * ** .:::* :: :
Mb3157c|M.bovis_AF2122/97 RGAAEKQDPLSG---LTDQERTLLGLLSEGLTNKQIADRMFLAEKTVKNY
Rv3133c|M.tuberculosis_H37Rv RGAAEKQDPLSG---LTDQERTLLGLLSEGLTNKQIADRMFLAEKTVKNY
Mvan_1646|M.vanbaalenii_PYR-1 LNRARHGDDLPG-TIVTSREEEVLKLIAEGYSTREIAGTLGISAKTVDRH
MSMEG_0856|M.smegmatis_MC2_155 LHRARQGGEPPD-SILTPREEEVLKLIAEGYSTREIAATLTISAKTVDRH
MMAR_4782|M.marinum_M AGEIRQRAAPMA-PVLSAREREVLHRIAKGQSIPAIAGELYVAPSTVKTH
MUL_0351|M.ulcerans_Agy99 AGEIRQRAAPMA-PVLSAREREVLHRIAKGQSIPAIAGELYVAPSTVKTH
Mflv_5014|M.gilvum_PYR-GCK --------WPGARQGLTQRESEILALVVNGLSNRAIAAKLIIGDETVKTH
TH_3184|M.thermoresistible__bu ARLQRDEFWPGARHGLTQRESEILSYVVNGLSNRGIAAKLVIGDETVKSH
MAB_1522|M.abscessus_ATCC_1997 RRAPAAVSGDAQ-DKLTARELDVLALIASGATNSEIAEQLVISEVTVKSH
MAV_5280|M.avium_104 RKAAPGPHGAAI-AELTTRERDVLTLIGKGLSNSEIADELCISGVTVKSH
MLBr_01286|M.leprae_Br4923 ADRLETRKLVER----AKGLLQVKQGMTEPEAFKWIQRAAMDRRTTMKRV
: : : . : * *:.
Mb3157c|M.bovis_AF2122/97 VSRLLAKLGMERRTQAAVFATELKRSRPPGDGP
Rv3133c|M.tuberculosis_H37Rv VSRLLAKLGMERRTQAAVFATELKRSRPPGDGP
Mvan_1646|M.vanbaalenii_PYR-1 RANLLAKLGLRDRLALTRYAIRVGLIEP-----
MSMEG_0856|M.smegmatis_MC2_155 RTNILAKLGLRDRLALTRYAIRAGLIEP-----
MMAR_4782|M.marinum_M VQRLYEKLGVSDRAAAVAEAMRQGLLD------
MUL_0351|M.ulcerans_Agy99 VQRLYEKLGVSDRAAAVAEAMRQGLLD------
Mflv_5014|M.gilvum_PYR-GCK LSSIYRKLGVSDRTAAVATALREGIFQ------
TH_3184|M.thermoresistible__bu LRSIYRKLGVSDRAAAVATALREGIYQ------
MAB_1522|M.abscessus_ATCC_1997 IGRIFVKLDLRDRAAAIVYAYDHGIVTPK----
MAV_5280|M.avium_104 IGRIFGKLDLRDRAAAIVYAYDNGIVAPR----
MLBr_01286|M.leprae_Br4923 AEVVLETLDTPKDT-------------------
: .*.