For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DVYEAVRSRRAVRGFTDEPVPQAVLERVLAAAAWAPSGSNLQPWHIYVVTGAPLAQMKKLATERVAAGDP WDEREYVMYPPVLKSPYHERRATFGRERYSALGIEREDWEARQRAAIANWDCFGAPAALFCYIDRDLGRP QWSDLGMYLQTVMLLLRAEGLHSCPQMAWSQVRSTVAQVLSPPDELMLFCGMSIGHEDTSVGYVPTGRSA LAETVTFVNG*
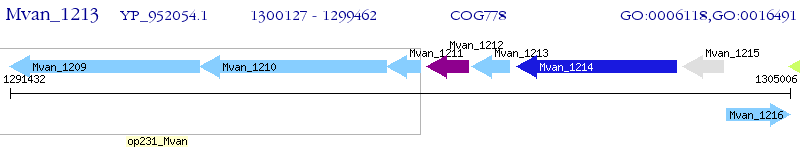
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_1213 | - | - | 100% (221) | nitroreductase |
| M. vanbaalenii PYR-1 | Mvan_5547 | - | 1e-10 | 50.98% (51) | nitroreductase |
| M. vanbaalenii PYR-1 | Mvan_4554 | - | 4e-07 | 46.00% (50) | nitroreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1262 | - | 1e-10 | 42.03% (69) | nitroreductase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2661c | - | 1e-71 | 57.99% (219) | putative nitroreductase |
| M. marinum M | MMAR_1739 | - | 2e-30 | 33.03% (221) | nitroreductase |
| M. avium 104 | MAV_0933 | - | 2e-08 | 40.00% (90) | nitroreductase family protein |
| M. smegmatis MC2 155 | MSMEG_3462 | - | 1e-100 | 79.82% (218) | oxidoreductase |
| M. thermoresistible (build 8) | TH_1618 | - | 3e-10 | 50.94% (53) | nitroreductase family protein |
| M. ulcerans Agy99 | MUL_1982 | - | 1e-17 | 34.35% (131) | nitroreductase |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_1213|M.vanbaalenii_PYR-1 -------MDVYEAVRSRRAVRGFTDEPVPQAVLERVLAAAAWAPSGSNLQ
MSMEG_3462|M.smegmatis_MC2_155 ------MSDVYEAVISRRAVRGFTDRPVPKAVLARVLSAAAWSPSSSNTQ
MAB_2661c|M.abscessus_ATCC_199 -------MDVYDAVASRRSVREFVDRPVSPATLTRVLTAAARAPSGGNLQ
MMAR_1739|M.marinum_M ------MYDLDETIRLRRSTRMFLPRPVPRRLVQEALELAMCAPSNSNVQ
MUL_1982|M.ulcerans_Agy99 --------------------------------------------------
Mflv_1262|M.gilvum_PYR-GCK MDMPHPTDNVWEVLSTARSIRRFTDEPVDSATLDRCLQAATWAPNGANAQ
TH_1618|M.thermoresistible__bu --MPHPTNDVWEVLSTARSIRRFTDEPVDDATLQRCLQAATWAPNGANAQ
MAV_0933|M.avium_104 ------------MMSTARTIRRFTDEPVDDATLARCLGAARWAPSGANAQ
Mvan_1213|M.vanbaalenii_PYR-1 PWHIYVVTGAPLAQMKKLATERVAAGDPWDEREYVMYPPVLKSPYHERRA
MSMEG_3462|M.smegmatis_MC2_155 PWNIYVVTGDALAALKKRATERVAMGVPWDEREYDMYPATMTSPYADRRA
MAB_2661c|M.abscessus_ATCC_199 PWHVYVLSGARLAELKERIARRVIAGDRGDALEVAMYPQPLDSPYRERLY
MMAR_1739|M.marinum_M PWHVVFTSGAARDRLVAAMLERARSEPPNVAR--------LPPAFTHLRS
MUL_1982|M.ulcerans_Agy99 ----------------------------------------MPPAFTHLRS
Mflv_1262|M.gilvum_PYR-GCK LWRFIVLDGPEQRAAVAQAAQ---------------------IALQSIES
TH_1618|M.thermoresistible__bu LWRFIVLDAPEQRAAVAEAAQ---------------------IALHSIES
MAV_0933|M.avium_104 GWRFVVLRSPEQRAVVAKAAA---------------------QALEVIEP
.
Mvan_1213|M.vanbaalenii_PYR-1 TFGRERYSALGIEREDWEARQRAAIANWDCFGAPAALFCYIDRDLGRPQW
MSMEG_3462|M.smegmatis_MC2_155 AFGKERYSALGIARDDWEARQRAAIANWDCFGAPAALFCYIDRGLGPAQW
MAB_2661c|M.abscessus_ATCC_199 DFGHRRYGALGIDHDDHDARAAVRAGNWACFGASTALFCYLDRDMPPPQW
MMAR_1739|M.marinum_M ALGAQVYGAMGIARDDTQARQTAVLRNWEFFRAPLGGIVSMHQELGLVDS
MUL_1982|M.ulcerans_Agy99 ALGAQVYGAMGIARDDTQARQTAVLRNWEFFRAPLGGIVSMHQELGLVDS
Mflv_1262|M.gilvum_PYR-GCK IYGMTRPAPEDDSRSARNNRATYELHDRAG-EFTSVLFVAYKNEFSSEFL
TH_1618|M.thermoresistible__bu IYGMSRPADDDRSRAARNNRATYELHDRAG-EYTSVLFTAFKNEFSSELL
MAV_0933|M.avium_104 VYGMSRPAPDDHSRRARTYRATYELHDRAG-EFTSVLFAQAHYPTASELL
* . . : * : . : .
Mvan_1213|M.vanbaalenii_PYR-1 SDLGMY--LQTVMLLLRAEGLHSCPQMAWSQVR--STVAQVLSPPDELML
MSMEG_3462|M.smegmatis_MC2_155 ADLGMY--LQTVMLLLRAEGLHSCPQMAWSQVR--TTVAEAVSPPPRLML
MAB_2661c|M.abscessus_ATCC_199 GDAGMY--LQTIMLLLRAEGLDSCPQIAWAEYH--RTVAEVIAPPPQRVL
MMAR_1739|M.marinum_M MGVGMF--LQNLMLALTARGLGTCVQVSIAGYP--EIVRKQVGIPSHLTI
MUL_1982|M.ulcerans_Agy99 MGVGMF--LQNLMLALTARGLGTCVQVSIAGYP--EIVRKQVGIPSHLTI
Mflv_1262|M.gilvum_PYR-GCK QGGSIYPALQNFYLAARAQGLGACITS-WASYGGEQALREAVGIPDEWFL
TH_1618|M.thermoresistible__bu QGGSIYPAMQNFYLAARAQGLGACFTS-WASYEGEKVLREAVGVPDDWFL
MAV_0933|M.avium_104 LGGSIFPAMQNFLLAARAQGLGACLTS-WASYGGEQLLREAVGVPEGWMI
. .:: :*.. * *.** :* : : : :. * :
Mvan_1213|M.vanbaalenii_PYR-1 FCGMSIGHEDT--SVGYVPTGRSALAETVTFVNG----------------
MSMEG_3462|M.smegmatis_MC2_155 FCGMSIGYEDP--SVDYVRTGRAPLKETVTFVQ-----------------
MAB_2661c|M.abscessus_ATCC_199 YCGMSIGYRNP--ETKAPEMPRAALSETVTFL------------------
MMAR_1739|M.marinum_M LCGLAIGYPDPDFAGNQLRVGRDDVAEHVVFLDD----------------
MUL_1982|M.ulcerans_Agy99 LCGLAIGYPDPDFAGNQLRVGRDDVAEHVVFLDD----------------
Mflv_1262|M.gilvum_PYR-GCK AGHVVVGWPRG----KHGPVRRRPLSDVVFRNHWDPERADITYGRGARPR
TH_1618|M.thermoresistible__bu AGHVVVGWPRG----RHGPVRRRPLGDVVFRNRWDPDRADITYGRGARPK
MAV_0933|M.avium_104 AGHIVVGWPKG----KHGPLRRRPLAEFVNLDRWDAPADGLLSGL-----
: :* * : : *
Mvan_1213|M.vanbaalenii_PYR-1 --
MSMEG_3462|M.smegmatis_MC2_155 --
MAB_2661c|M.abscessus_ATCC_199 --
MMAR_1739|M.marinum_M --
MUL_1982|M.ulcerans_Agy99 --
Mflv_1262|M.gilvum_PYR-GCK GK
TH_1618|M.thermoresistible__bu RR
MAV_0933|M.avium_104 --