For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LPPAFTHLRSALGAQVYGAMGIARDDTQARQTAVLRNWEFFRAPLGGIVSMHQELGLVDSMGVGMFLQNL MLALTARGLGTCVQVSIAGYPEIVRKQVGIPSHLTILCGLAIGYPDPDFAGNQLRVGRDDVAEHVVFLDD
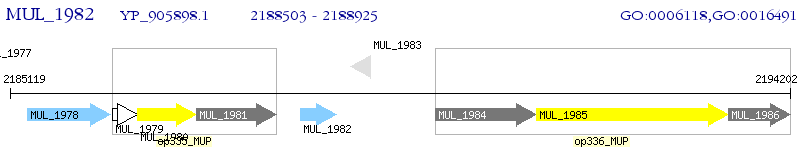
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_1982 | - | - | 100% (140) | nitroreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2661c | - | 8e-14 | 32.54% (126) | putative nitroreductase |
| M. marinum M | MMAR_1739 | - | 4e-78 | 100.00% (140) | nitroreductase |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2354 | - | 8e-46 | 61.31% (137) | oxidoreductase |
| M. thermoresistible (build 8) | TH_0777 | - | 2e-05 | 40.38% (52) | PROBABLE F420 BIOSYNTHESIS PROTEIN FBIB |
| M. vanbaalenii PYR-1 | Mvan_1213 | - | 7e-18 | 34.35% (131) | nitroreductase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_1982|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1739|M.marinum_M --------------------------------------------------
MSMEG_2354|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2661c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_1213|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0777|M.thermoresistible__bu VSTAGGAGQTGSGDHGTAAAIELFPVTGLPQFCPGDDLTAAVAAAAPWLR
MUL_1982|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1739|M.marinum_M --------------------------------------------------
MSMEG_2354|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2661c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_1213|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0777|M.thermoresistible__bu DNDVVVVTSKVVSKCEGRIIAAPADPEQRDALRRKLIDEEAVRVLARKGR
MUL_1982|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1739|M.marinum_M --------------------------------------------------
MSMEG_2354|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2661c|M.abscessus_ATCC_199 --------------------------------------------------
Mvan_1213|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_0777|M.thermoresistible__bu TLITENALGLVQAAAGVDGSNVAATELALLPVDPDASAAALRAGLRQRLG
MUL_1982|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1739|M.marinum_M ------------------------------MYDLDETIRLRRSTRMFLP-
MSMEG_2354|M.smegmatis_MC2_155 -------------------------------MELAEAVRARRSTRKFLPD
MAB_2661c|M.abscessus_ATCC_199 -------------------------------MDVYDAVASRRSVREFVD-
Mvan_1213|M.vanbaalenii_PYR-1 -------------------------------MDVYEAVRSRRAVRGFTD-
TH_0777|M.thermoresistible__bu VDVGVIITDTMGRAWRTGQTDFAIGAAGLTVLHGYAGSHDRHGNELFVTE
MUL_1982|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1739|M.marinum_M ------------------RPVPRRLVQEALELAMCAPSNSNVQP------
MSMEG_2354|M.smegmatis_MC2_155 ------------------KPVPHELIEEALTLAMRAPSNSNVQP------
MAB_2661c|M.abscessus_ATCC_199 ------------------RPVSPATLTRVLTAAARAPSGGNLQP------
Mvan_1213|M.vanbaalenii_PYR-1 ------------------EPVPQAVLERVLAAAAWAPSGSNLQP------
TH_0777|M.thermoresistible__bu VAVADEIAAAADLVKGKLTDVPVAVVRGLTLRDDGSTARTLVRPGQDDLF
MUL_1982|M.ulcerans_Agy99 --------------------------------------------------
MMAR_1739|M.marinum_M ---------------------WHVVFTSGAARDRLVAAMLERARSEPPNV
MSMEG_2354|M.smegmatis_MC2_155 ---------------------WRVYFASGERRERLVAALTAEVRARRPTS
MAB_2661c|M.abscessus_ATCC_199 ---------------------WHVYVLSGARLAELKERIARRVIAGDRGD
Mvan_1213|M.vanbaalenii_PYR-1 ---------------------WHIYVVTGAPLAQMKKLATERVAAGDPWD
TH_0777|M.thermoresistible__bu WLGTEEAIRLGRSQAQLLRRSVRRFDSEPVPADLIEAAVAEALTAPAPHH
MUL_1982|M.ulcerans_Agy99 ----------MPPAFTHLRSALGAQVYGAMGIARDDTQARQTAVLRNWEF
MMAR_1739|M.marinum_M AR--------LPPAFTHLRSALGAQVYGAMGIARDDTQARQTAVLRNWEF
MSMEG_2354|M.smegmatis_MC2_155 LG--------LPDAFQHLRSQLGALVYGSMGIARADTEARWAAQLRNWEF
MAB_2661c|M.abscessus_ATCC_199 ALEVAMYPQPLDSPYRERLYDFGHRRYGALGIDHDDHDARAAVRAGNWAC
Mvan_1213|M.vanbaalenii_PYR-1 EREYVMYPPVLKSPYHERRATFGRERYSALGIEREDWEARQRAAIANWDC
TH_0777|M.thermoresistible__bu TRPVRFVWLQNWTARTRLLDRMKDRWRADLAGDGASPEAVERRVGRGQIL
. . : . :. . :* .
MUL_1982|M.ulcerans_Agy99 FRAPLGGIVSMHQELGLVD-------------SMGVGMFLQNLMLALTAR
MMAR_1739|M.marinum_M FRAPLGGIVSMHQELGLVD-------------SMGVGMFLQNLMLALTAR
MSMEG_2354|M.smegmatis_MC2_155 FGAPLAGVVCMHRDLGLVD-------------SVGVGMFLQTFLLALTER
MAB_2661c|M.abscessus_ATCC_199 FGASTALFCYLDRDMPPPQ-------------WGDAGMYLQTIMLLLRAE
Mvan_1213|M.vanbaalenii_PYR-1 FGAPAALFCYIDRDLGRPQ-------------WSDLGMYLQTVMLLLRAE
TH_0777|M.thermoresistible__bu YDAPELIVPFLVADGAHHYPDARRAAAEHTMFTVAGGAAVQALLVALAVR
: *. . : : * :* .:: * .
MUL_1982|M.ulcerans_Agy99 GLGTCVQVSIAGYPEIVRKQVGIPSHLTILCGLAIGYPDPDFAGNQLRVG
MMAR_1739|M.marinum_M GLGTCVQVSIAGYPEIVRKQVGIPSHLTILCGLAIGYPDPDFAGNQLRVG
MSMEG_2354|M.smegmatis_MC2_155 GIGSCVQVSVAMCADVLRAELDIPDELTVLCGMSIGYADPAFPANHLDIP
MAB_2661c|M.abscessus_ATCC_199 GLDSCPQIAWAEYHRTVAEVIAPPPQRVLYCGMSIGYRNP--ETKAPEMP
Mvan_1213|M.vanbaalenii_PYR-1 GLHSCPQMAWSQVRSTVAQVLSPPDELMLFCGMSIGHEDT--SVGYVPTG
TH_0777|M.thermoresistible__bu EIGSCWVGSTIFAPEVVRTELGLPDDWEPLGAVAIGYPAEP----AGPRD
: :* : : : * . .::**:
MUL_1982|M.ulcerans_Agy99 RDDVAEHVVFLDD
MMAR_1739|M.marinum_M RDDVAEHVVFLDD
MSMEG_2354|M.smegmatis_MC2_155 RNPVEQNVTFYS-
MAB_2661c|M.abscessus_ATCC_199 RAALSETVTFL--
Mvan_1213|M.vanbaalenii_PYR-1 RSALAETVTFVNG
TH_0777|M.thermoresistible__bu PAPTEGLLVFR--
:.*